7YG4
 
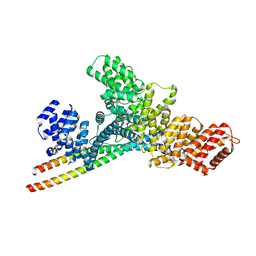 | | Structure of WTAP-VIRMA in the m6A writer complex | | Descriptor: | Pre-mRNA-splicing regulator WTAP, Protein virilizer homolog | | Authors: | Yan, X.H, Guan, Z.Y, Tang, C, Yin, P. | | Deposit date: | 2022-07-11 | | Release date: | 2022-11-23 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | AI-empowered integrative structural characterization of m 6 A methyltransferase complex.
Cell Res., 32, 2022
|
|
2MP0
 
 | | Protein Phosphorylation upon a Fleeting Encounter | | Descriptor: | Glucose-specific phosphotransferase enzyme IIA component, PHOSPHITE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Xing, Q, Yang, J, Huang, P, Zhang, W, Tang, C. | | Deposit date: | 2014-05-08 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Visualizing an ultra-weak protein-protein interaction in phosphorylation signaling.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2KI0
 
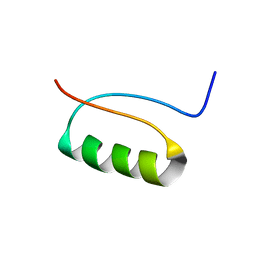 | | NMR Structure of a de novo designed beta alpha beta | | Descriptor: | DS119 | | Authors: | Liang, H, Chen, H, Fan, K, Wei, P, Guo, X, Jin, C, Zeng, C, Tang, C, Lai, L. | | Deposit date: | 2009-04-18 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | De novo design of a beta alpha beta motif.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
5YMY
 
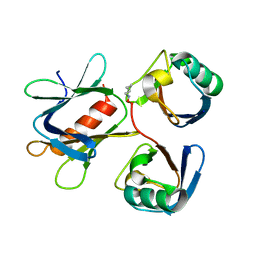 | | The structure of the complex between Rpn13 and K48-diUb | | Descriptor: | Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Liu, Z, Dong, X, Gong, Z, Yi, H.W, Liu, K, Yang, J, Zhang, W.P, Tang, C. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of K48-linked Ub chain by proteasomal receptor Rpn13.
Cell Discov, 5, 2019
|
|
5YZ9
 
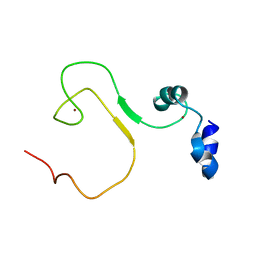 | | zinc finger domain of METTL3-METTL14 N6-methyladenosine methyltransferase | | Descriptor: | N6-adenosine-methyltransferase catalytic subunit, ZINC ION | | Authors: | Dong, X, Tang, C, Gong, Z, Yin, P, Huang, J.B. | | Deposit date: | 2017-12-13 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of the RNA recognition domain of METTL3-METTL14 N6-methyladenosine methyltransferase.
Protein Cell, 10, 2019
|
|
6BRK
 
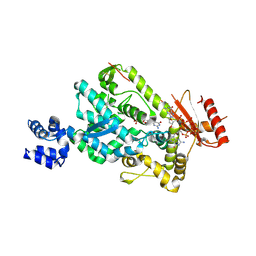 | | The SAM domain of mouse SAMHD1 is critical for its activation and regulation | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION | | Authors: | Buzovetsky, O, Tang, C, Knecht, K.M, Antonucci, J.M, Wu, L, Ji, X, Xiong, Y. | | Deposit date: | 2017-11-30 | | Release date: | 2018-02-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The SAM domain of mouse SAMHD1 is critical for its activation and regulation.
Nat Commun, 9, 2018
|
|
2JC5
 
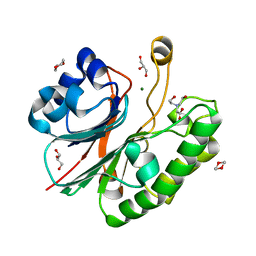 | | Apurinic Apyrimidinic (AP) endonuclease (NApe) from Neisseria Meningitidis | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, BICINE, EXODEOXYRIBONUCLEASE, ... | | Authors: | Carpenter, E.P, Corbett, A, Thomson, H, Adacha, J, Jensen, K, Bergeron, J, Kasampalidis, I, Exley, R, Winterbotham, M, Tang, C, Baldwin, G.S, Freemont, P. | | Deposit date: | 2006-12-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ap Endonuclease Paralogues with Distinct Activities in DNA Repair and Bacterial Pathogenesis.
Embo J., 26, 2007
|
|
2JC4
 
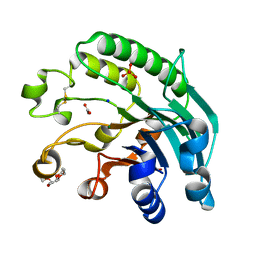 | | 3'-5' exonuclease (NExo) from Neisseria Meningitidis | | Descriptor: | ACETATE ION, DIHYDROGENPHOSPHATE ION, EXODEOXYRIBONUCLEASE III, ... | | Authors: | Carpenter, E.P, Corbett, A, Thomson, H, Adacha, J, Jensen, K, Bergeron, J, Kasampalidis, I, Exley, R, Winterbotham, M, Tang, C, Baldwin, G, Freemont, P. | | Deposit date: | 2006-12-19 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ap Endonuclease Paralogues with Distinct Activities in DNA Repair and Bacterial Pathogenesis.
Embo J., 26, 2007
|
|
2JPR
 
 | | Joint refinement of the HIV-1 CA-NTD in complex with the assembly inhibitor CAP-1 | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-{2-[({5-[(dimethylamino)methyl]-2-furyl}methyl)thio]ethyl}urea, Gag-Pol polyprotein | | Authors: | Kelly, B.N, Kyere, S, Kinde, I, Tang, C, Howard, B.R, Robinson, H, Sundquist, W.I, Summers, M.F, Hill, C.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein
J.Mol.Biol., 373, 2007
|
|
6KBH
 
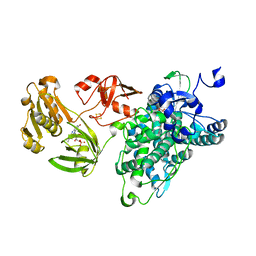 | | Crystal structure of an intact type IV self-sufficient cytochrome P450 monooxygenase | | Descriptor: | Cytochrome P450 monooxygenase, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gong, R, Wu, L.J, Zhang, Y, Liu, Z, Dou, S, Zhang, R.G, Xu, J.H, Tang, C, Zhou, J.H. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an intact type IV self-sufficient cytochrome P450 monooxygenase
To Be Published
|
|
2N2K
 
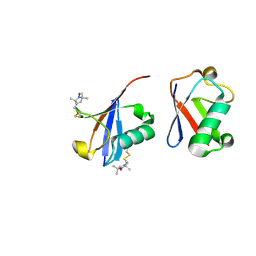 | | Ensemble structure of the closed state of Lys63-linked diubiquitin in the absence of a ligand | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, ubiquitin | | Authors: | Liu, Z, Gong, Z, Tang, C. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-08 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Lys63-linked ubiquitin chain adopts multiple conformational states for specific target recognition.
Elife, 4, 2015
|
|
6KOX
 
 | |
6ZV6
 
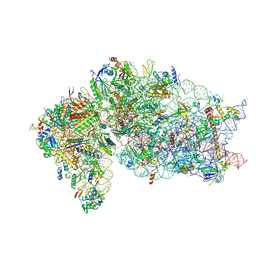 | | Human RIO1(kd)-StHA late pre-40S particle, structural state B (post 18S rRNA cleavage) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Plassart, L, Shayan, R, Plisson-Chastang, C. | | Deposit date: | 2020-07-24 | | Release date: | 2021-05-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The final step of 40S ribosomal subunit maturation is controlled by a dual key lock.
Elife, 10, 2021
|
|
6ZUO
 
 | | Human RIO1(kd)-StHA late pre-40S particle, structural state A (pre 18S rRNA cleavage) | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Plassart, L, Shayan, R, Plisson-Chastang, C. | | Deposit date: | 2020-07-23 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The final step of 40S ribosomal subunit maturation is controlled by a dual key lock.
Elife, 10, 2021
|
|
6Y7C
 
 | | Early cytoplasmic yeast pre-40S particle (purified with Tsr1 as bait) | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Plassart, L, Plisson-Chastang, C. | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Good Vibrations: Structural Remodeling of Maturing Yeast Pre-40S Ribosomal Particles Followed by Cryo-Electron Microscopy.
Molecules, 25, 2020
|
|
6RBE
 
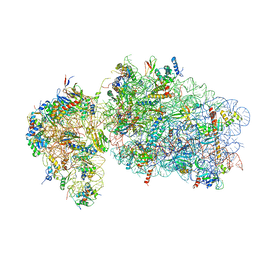 | | State 2 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
6RBD
 
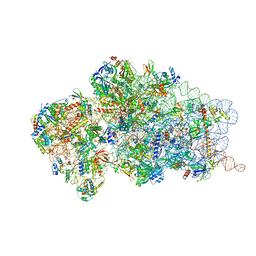 | | State 1 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
6LR3
 
 | | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum | | Descriptor: | Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Su, Z.M, Tian, X.Y, Li, H.J, Wei, Z.M, Chen, L.F, Ren, H.X, Peng, W.F, Tang, C.T. | | Deposit date: | 2020-01-15 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum.
Biochem.J., 477, 2020
|
|
5DIJ
 
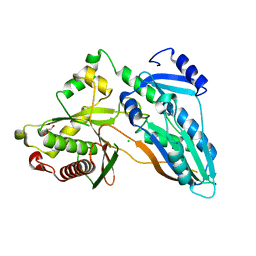 | | The crystal structure of CT | | Descriptor: | CHLORIDE ION, GLYCEROL, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-09-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
5DLK
 
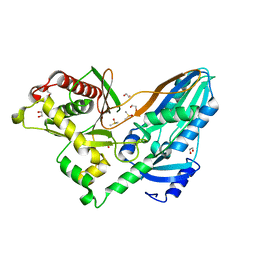 | | The crystal structure of CT mutant | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-09-06 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
6PM9
 
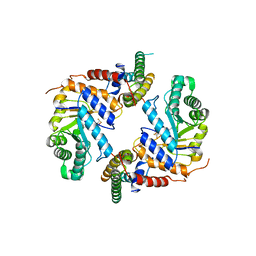 | | Crystal structure of the core catalytic domain of human O-GlcNAcase bound to MK-8719 | | Descriptor: | (3aR,5S,6S,7R,7aR)-5-(difluoromethyl)-2-(ethylamino)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, O-GlcNAcase TIM-barrel domain, O-GlcNAcase stalk domain | | Authors: | Klein, D.J, Selnick, H.G, Duffy, J.L, McEachern, E.J. | | Deposit date: | 2019-07-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of MK-8719, a Potent O-GlcNAcase Inhibitor as a Potential Treatment for Tauopathies.
J.Med.Chem., 62, 2019
|
|
5EGF
 
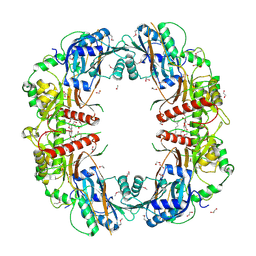 | | The crystal structure of SeMet-CT | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-10-27 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
5EJD
 
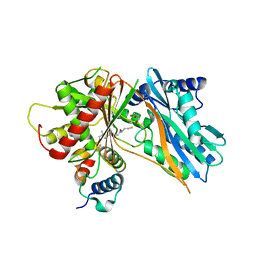 | | The crystal structure of holo T3CT | | Descriptor: | 4'-PHOSPHOPANTETHEINE, GLYCEROL, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
6LKV
 
 | | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum | | Descriptor: | CHLORIDE ION, Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Su, Z.M, Tian, X.Y, Li, H.J, Wei, Z.M, Chen, L.F, Ren, H.X, Peng, W.F, Tang, C.T. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum.
Biochem.J., 477, 2020
|
|
6LKW
 
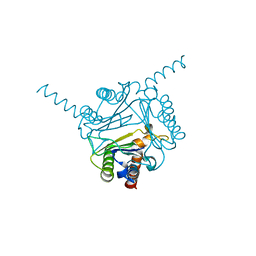 | | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum | | Descriptor: | CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Su, Z.M, Tian, X.Y, Li, H.J, Wei, Z.M, Chen, L.F, Ren, H.X, Peng, W.F, Tang, C.T. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum.
Biochem.J., 477, 2020
|
|
