4EUO
 
 | | Structure of Atu4243-GABA sensor | | Descriptor: | ABC transporter, substrate binding protein (Polyamine), GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Morera, S, Planamente, S. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural basis for selective GABA binding in bacterial pathogens.
Mol.Microbiol., 86, 2012
|
|
4EQ7
 
 | | Structure of Atu4243-GABA receptor | | Descriptor: | ABC transporter, substrate binding protein (Polyamine), GLYCEROL, ... | | Authors: | Morera, S, Planamente, S. | | Deposit date: | 2012-04-18 | | Release date: | 2012-11-21 | | Last modified: | 2012-12-19 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for selective GABA binding in bacterial pathogens.
Mol.Microbiol., 86, 2012
|
|
5DMV
 
 | | Polo-box domain of Mouse Polo-like kinase 1 complexed with Emi2 (146-177) | | Descriptor: | F-box only protein 43, NICKEL (II) ION, Serine/threonine-protein kinase PLK1 | | Authors: | Namgoong, S. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural basis for recognition of Emi2 by Polo-like kinase 1 and development of peptidomimetics blocking oocyte maturation and fertilization.
Sci Rep, 5, 2015
|
|
5DNJ
 
 | | Mouse Polo-box domain and Peptide analog 702 | | Descriptor: | Serine/threonine-protein kinase PLK1, peptide 707-56A-SER-TPO-NH2 | | Authors: | Namgoong, S, Han, Y.H. | | Deposit date: | 2015-09-10 | | Release date: | 2016-02-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of Emi2 by Polo-like kinase 1 and development of peptidomimetics blocking oocyte maturation and fertilization.
Sci Rep, 5, 2015
|
|
5TBW
 
 | | Crystal structure of chlorolissoclimide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Konst, Z.A, Szklarski, A.R, Pellegrino, S, Michalak, S.E, Meyer, M, Zanette, C, Cencic, R, Nam, S, Horne, D.A, Pelletier, J, Mobley, D.L, Yusupova, G, Yusupov, M, Vanderwal, C.D. | | Deposit date: | 2016-09-13 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis facilitates an understanding of the structural basis for translation inhibition by the lissoclimides.
Nat Chem, 9, 2017
|
|
3M3N
 
 | | Structure of a Longitudinal Actin Dimer Assembled by Tandem W Domains | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rebowski, G, Namgoong, S, Dominguez, R. | | Deposit date: | 2010-03-09 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Structure of a longitudinal actin dimer assembled by tandem w domains: implications for actin filament nucleation.
J.Mol.Biol., 403, 2010
|
|
4UQX
 
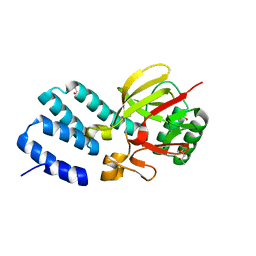 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, HSIE1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
J.Biol.Chem., 289, 2014
|
|
4UQW
 
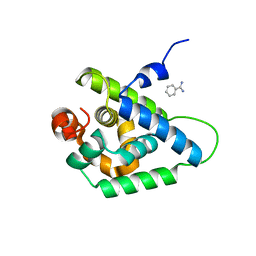 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | BENZAMIDINE, PROTEIN CLPV1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
J.Biol.Chem., 289, 2014
|
|
4UQY
 
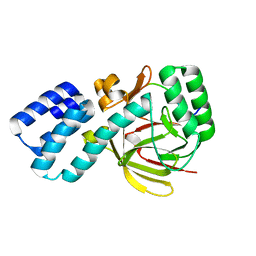 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | HSIB1, HSIE1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
J.Biol.Chem., 289, 2014
|
|
4UQZ
 
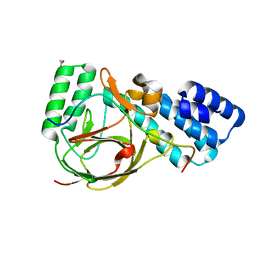 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | ACETATE ION, HSIB1, HSIE1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
RNA, 20, 2014
|
|
8SG6
 
 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant Reduced with 20mM TCEP | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL | | Authors: | Tran, N, McLeod, M.J, Barwell, S, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2023-04-11 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
7CK5
 
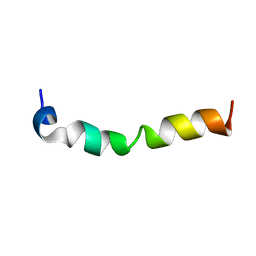 | | Solution structure of 28 amino acid polypeptide (354-381) in Plantago asiatica mosaic virus replicase bound to SDS micelle | | Descriptor: | PlAMV replicase peptide from RNA-dependent RNA polymerase | | Authors: | Komatsu, K, Sasaki, N, Yoshida, T, Suzuki, K, Masujima, Y, Hashimoto, M, Watanabe, S, Tochio, N, Kigawa, T, Yamaji, Y, Oshima, K, Namba, S, Nelson, R, Arie, T. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of a Proline-Kinked Amphipathic alpha-Helix Downstream from the Methyltransferase Domain of a Potexvirus Replicase and Its Role in Virus Replication and Perinuclear Complex Formation.
J.Virol., 95, 2021
|
|
5NXF
 
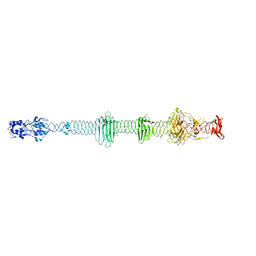 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, residues 795 to 1289, at 1.9 Angstrom. | | Descriptor: | ACETATE ION, GLYCEROL, Long-tail fiber proximal subunit, ... | | Authors: | Namura, M, van Raaij, M.J, Kanamaru, S. | | Deposit date: | 2017-05-10 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
5NXH
 
 | |
6Z2I
 
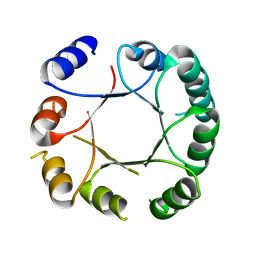 | | Crystal structure of DeNovoTIM6, a de novo designed TIM barrel | | Descriptor: | de novo designed TIM barrel DeNovoTIM6 | | Authors: | Romero-Romero, S, Kordes, S, Shanmugaratnam, S, Rodriguez-Romero, A, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-05-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
5N8N
 
 | | Contracted sheath of a Pseudomonas aeruginosa type six secretion system consisting of TssB1 and TssC1 | | Descriptor: | EvpB family type VI secretion protein, Type VI secretion protein, family | | Authors: | Salih, O, He, S, Stach, L, Macdonald, J.T, Planamente, S, Manoli, E, Scheres, S, Filloux, A, Freemont, P.S. | | Deposit date: | 2017-02-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Atomic Structure of Type VI Contractile Sheath from Pseudomonas aeruginosa.
Structure, 26, 2018
|
|
5NOP
 
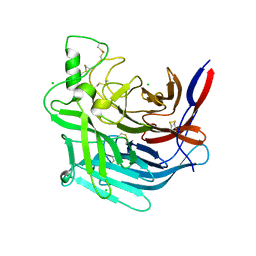 | | Structure of Mojiang virus attachment glycoprotein | | Descriptor: | Attachment glycoprotein, CHLORIDE ION | | Authors: | Rissanen, I.R, Ahmed, A.A, Beaty, S, Azarm, K, Hong, P, Nambulli, S, Duprex, P.W, Lee, B, Bowden, T.A. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Idiosyncratic Mojiang virus attachment glycoprotein directs a host-cell entry pathway distinct from genetically related henipaviruses.
Nat Commun, 8, 2017
|
|
7OYY
 
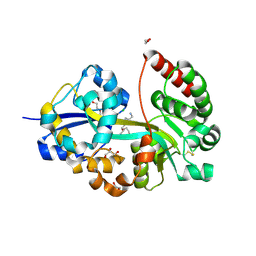 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutation S247D in complex with spermidine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kroeger, P, Shanmugaratnam, S, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
6YQY
 
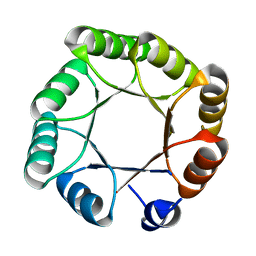 | | Crystal structure of sTIM11noCys, a de novo designed TIM barrel | | Descriptor: | de novo designed TIM barrel sTIM11noCys | | Authors: | Romero-Romero, S, Wiese, G.J, Kordes, S, Shanmugaratnam, S, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-04-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
6YQX
 
 | | Crystal structure of DeNovoTIM13, a de novo designed TIM barrel | | Descriptor: | CHLORIDE ION, GLYCEROL, de novo designed TIM barrel DeNovoTIM13 | | Authors: | Romero-Romero, S, Kordes, S, Shanmugaratnam, S, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-04-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.638 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
3J2M
 
 | | The X-ray structure of the gp15 hexamer and the model of the gp18 protein fitted into the cryo-EM reconstruction of the extended T4 tail | | Descriptor: | Tail connector protein Gp15, Tail sheath protein Gp18 | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-09 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
3J2N
 
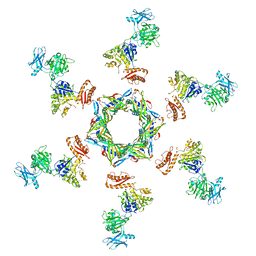 | | The X-ray structure of the gp15 hexamer and the model of the gp18 protein fitted into the cryo-EM reconstruction of the contracted T4 tail | | Descriptor: | Tail connector protein Gp15, Tail sheath protein Gp18 | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-10 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
3J2O
 
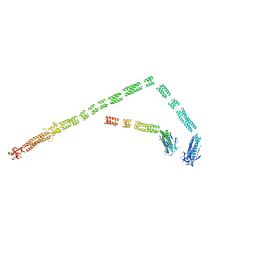 | | Model of the bacteriophage T4 fibritin based on the cryo-EM reconstruction of the contracted T4 tail containing the phage collar and whiskers | | Descriptor: | Fibritin | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A.A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-12 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
5YJI
 
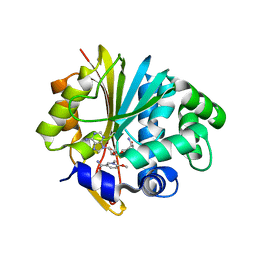 | | Co-crystal structure of Mouse Nicotinamide N-methyltransferase (NNMT) with small molecule analog of Nicotinamide | | Descriptor: | 6-methoxy-1-methyl-2H-pyridine-3-carboxamide, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Birudukota, S, Swaminathan, S, Thakur, M.K, Parveen, R, Kandan, S, Hallur, M.S, Rajagopal, S, Ruf, S, Dhakshinamoorthy, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A small molecule inhibitor of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 8, 2018
|
|
5YJF
 
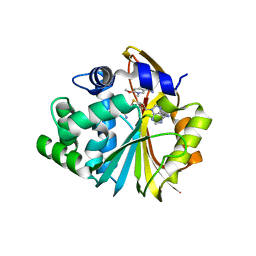 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with small molecule analog of Nicotinamide | | Descriptor: | 6-methoxy-1-methyl-2H-pyridine-3-carboxamide, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Swaminathan, S, Birudukota, S, Thakur, M.K, Parveen, R, Kandan, S, Hallur, M.S, Rajagopal, S, Ruf, S, Dhakshinamoorthy, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | A small molecule inhibitor of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 8, 2018
|
|
