4XJ4
 
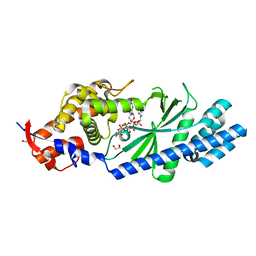 | | Crystal structure of Vibrio cholerae DncV 3'-deoxy ATP bound form | | Descriptor: | 1,2-ETHANEDIOL, 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-GMP synthase, ... | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
5EGE
 
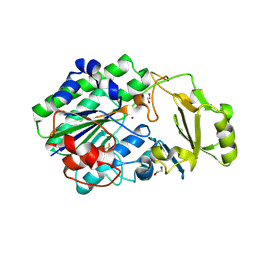 | | Structure of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Morita, J, Kano, K, Kato, K, Takita, H, Ishitani, R, Nishimasu, H, Nureki, O, Aoki, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-09 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase
Sci Rep, 6, 2016
|
|
4XJ5
 
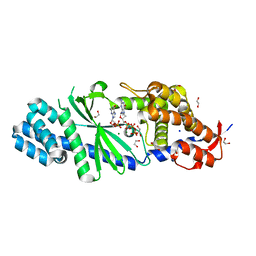 | | Crystal structure of Vibrio cholerae DncV 3'-deoxy GTP bound form | | Descriptor: | 1,2-ETHANEDIOL, 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, Cyclic AMP-GMP synthase, ... | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
5F1C
 
 | | Crystal structure of an invertebrate P2X receptor from the Gulf Coast tick in the presence of ATP and Zn2+ ion at 2.9 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Putative uncharacterized protein, ... | | Authors: | Kasuya, G, Hattori, M, Ishitani, R, Nureki, O. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into Divalent Cation Modulations of ATP-Gated P2X Receptor Channels
Cell Rep, 14, 2016
|
|
3G5R
 
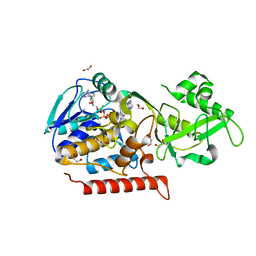 | | Crystal structure of Thermus thermophilus TrmFO in complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5EGH
 
 | | Structure of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase in complex with phosphocholine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Morita, J, Kano, K, Kato, K, Takita, H, Ishitani, R, Nishimasu, H, Nureki, O, Aoki, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase
Sci Rep, 6, 2016
|
|
3G5Q
 
 | | Crystal structure of Thermus thermophilus TrmFO | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate--tRNA-(uracil-5-)-methyltransferase trmFO | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G5S
 
 | | Crystal structure of Thermus thermophilus TrmFO in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, ... | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5GUH
 
 | | Crystal structure of silkworm PIWI-clade Argonaute Siwi bound to piRNA | | Descriptor: | MAGNESIUM ION, PIWI, RNA (28-MER) | | Authors: | Matsumoto, N, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Silkworm PIWI-Clade Argonaute Siwi Bound to piRNA
Cell, 167, 2016
|
|
3OQJ
 
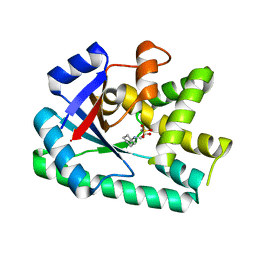 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC in complex with CAPSO | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4P79
 
 | | Crystal structure of mouse claudin-15 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Claudin-15 | | Authors: | Suzuki, H, Nishizawa, T, Tani, K, Yamazaki, Y, Tamura, A, Ishitani, R, Dohmae, N, Tsukita, S, Nureki, O, Fujiyoshi, Y. | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a claudin provides insight into the architecture of tight junctions.
Science, 344, 2014
|
|
3OQI
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC in complex with CHES | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OQH
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC | | Descriptor: | GLYCEROL, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4U4V
 
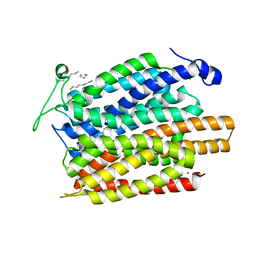 | | Structure of a nitrate/nitrite antiporter NarK in apo inward-open state | | Descriptor: | NICKEL (II) ION, Nitrate/nitrite transporter NarK, OLEIC ACID | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
4U4W
 
 | | Structure of a nitrate/nitrite antiporter NarK in nitrate-bound occluded state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, NITRATE ION, Nitrate/nitrite transporter NarK, ... | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
5Y78
 
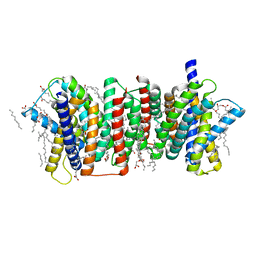 | | Crystal structure of the triose-phosphate/phosphate translocator in complex with inorganic phosphate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PHOSPHATE ION, Putative hexose phosphate translocator | | Authors: | Lee, Y, Nishizawa, T, Takemoto, M, Kumazaki, K, Yamashita, K, Hirata, K, Minoda, A, Nagatoishi, S, Tsumoto, K, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity
Nat Plants, 3, 2017
|
|
5Y79
 
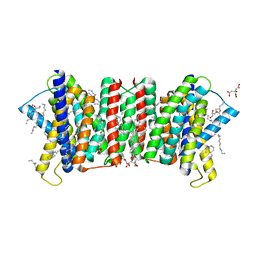 | | Crystal structure of the triose-phosphate/phosphate translocator in complex with 3-phosphoglycerate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-PHOSPHOGLYCERIC ACID, CITRATE ANION, ... | | Authors: | Lee, Y, Nishizawa, T, Takemoto, M, Kumazaki, K, Yamashita, K, Hirata, K, Minoda, A, Nagatoishi, S, Tsumoto, K, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity
Nat Plants, 3, 2017
|
|
6AI6
 
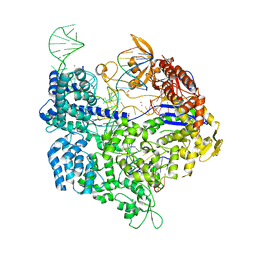 | | Crystal structure of SpCas9-NG | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, DNA (28-MER), ... | | Authors: | Nishimasu, H, Hirano, S, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-21 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineered CRISPR-Cas9 nuclease with expanded targeting space
Science, 361, 2018
|
|
6AEK
 
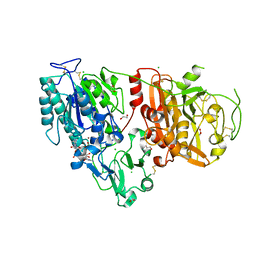 | | Crystal structure of ENPP1 in complex with pApG | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kato, K, Nishimasu, H, Hirano, S, Hirano, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-05 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1.
Nat Commun, 9, 2018
|
|
6AEL
 
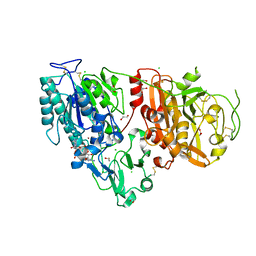 | | Crystal structure of ENPP1 in complex with 3'3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, ... | | Authors: | Kato, K, Nishimasu, H, Hirano, S, Hirano, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-05 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1.
Nat Commun, 9, 2018
|
|
5GIJ
 
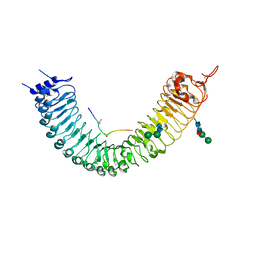 | | Crystal structure of TDR-TDIF complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat receptor-like protein kinase TDR, ... | | Authors: | Morita, J, Kato, K, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the plant receptor-like kinase TDR in complex with the TDIF peptide
Nat Commun, 7, 2016
|
|
5HRT
 
 | | Crystal structure of mouse autotaxin in complex with a DNA aptamer | | Descriptor: | CALCIUM ION, CHLORIDE ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Kato, K, Nishimasu, H, Morita, J, Ishitani, R, Nureki, O. | | Deposit date: | 2016-01-24 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural basis for specific inhibition of Autotaxin by a DNA aptamer
Nat.Struct.Mol.Biol., 23, 2016
|
|
5X2G
 
 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5X2H
 
 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5XH7
 
 | | Crystal structure of the Acidaminococcus sp. BV3L6 Cpf1 RR variant in complex with crRNA and target DNA (TCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CRISPR-associated endonuclease Cpf1, ... | | Authors: | Nishimasu, H, Yamano, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Mol. Cell, 67, 2017
|
|
