6LVP
 
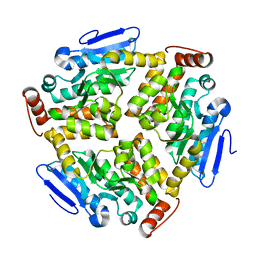 | |
2QMQ
 
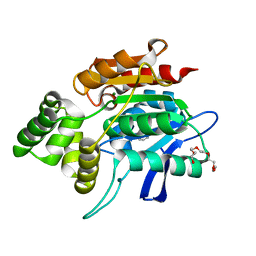 | |
7EHK
 
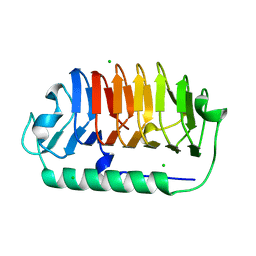 | | Crystal structure of C107S mutant of FfIBP | | Descriptor: | CHLORIDE ION, Ice-binding protein | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2021-03-29 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Importance of rigidity of ice-binding protein (FfIBP) for hyperthermal hysteresis activity and microbial survival.
Int.J.Biol.Macromol., 204, 2022
|
|
3SOV
 
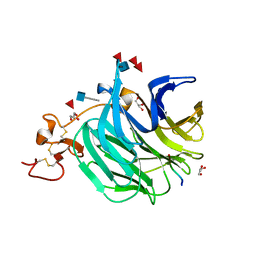 | | The structure of a beta propeller domain in complex with peptide S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Low-density lipoprotein receptor-related protein 6, ... | | Authors: | Wang, W, Bourhis, E, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
7XRI
 
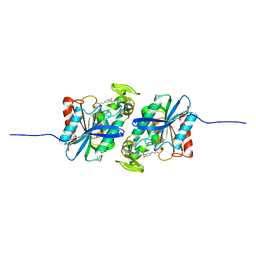 | | Feruloyl esterase mutant -S106A | | Descriptor: | Cinnamoyl esterase, ethyl (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate | | Authors: | Hwang, J.S, Lee, J.H, Do, H, Lee, C.W. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
2Q99
 
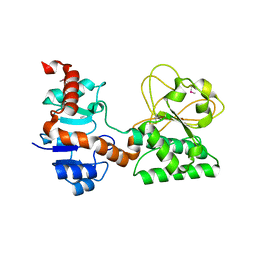 | |
6J10
 
 | | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly | | Descriptor: | 6-cyclohexyl-4-methyl-1-oxidanyl-pyridin-2-one, Capsid protein | | Authors: | Park, S, Jin, M.S, Cho, Y, Kang, J, Kim, S, Park, M, Park, H, Kim, J, Park, S, Hwang, J, Kim, Y, Kim, Y.J. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly.
Nat Commun, 10, 2019
|
|
7BXZ
 
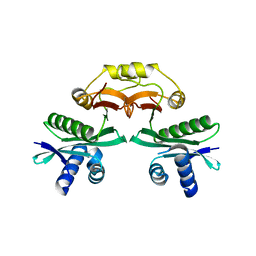 | |
3I0Q
 
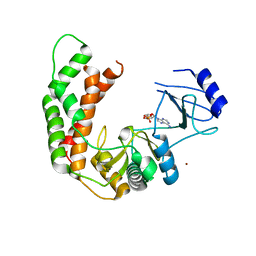 | | Crystal Structure of the AMP-bound complex of Spectinomycin Phosphotransferase, APH(9)-Ia | | Descriptor: | ADENOSINE MONOPHOSPHATE, NICKEL (II) ION, Spectinomycin phosphotransferase | | Authors: | Berghuis, A.M, Fong, D.H, Lemke, C.T, Hwang, J.-Y, Xiong, B. | | Deposit date: | 2009-06-25 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila.
J.Biol.Chem., 285, 2010
|
|
8G8A
 
 | | Crystal structure of DH1317.8 Fab in complex with HIV proximal MPER peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1317.8 heavy chain, DH1317.8 light chain, ... | | Authors: | Janus, B.M, Astavans, A, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
8G8C
 
 | | Crystal structure of DH1322.1 Fab in complex with HIV proximal MPER peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1322.1 heavy chain, DH1322.1 light chain, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
8G8D
 
 | | Crystal structure of DH1346 Fab in complex with HIV proximal MPER peptide | | Descriptor: | DH1346 heavy chain, DH1346 light chain, FLUORIDE ION, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
8K55
 
 | |
8K57
 
 | |
2I0X
 
 | | Hypothetical protein PF1117 from Pyrococcus furiosus | | Descriptor: | Hypothetical protein PF1117 | | Authors: | Chen, L.Q, Fu, Z.-Q, Liu, Z.-J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Hypothetical Protein Pf1117 from Pyrococcus furiosus
To be Published
|
|
7VPF
 
 | |
7XJT
 
 | |
7X99
 
 | |
3P0Y
 
 | | anti-EGFR/HER3 Fab DL11 in complex with domain III of EGFR extracellular region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2010-09-29 | | Release date: | 2011-10-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A two-in-one antibody against HER3 and EGFR has superior inhibitory activity compared with monospecific antibodies.
Cancer Cell, 20, 2011
|
|
3P0V
 
 | | anti-EGFR/HER3 Fab DL11 alone | | Descriptor: | CALCIUM ION, Fab DL11 heavy chain, Fab DL11 light chain | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2010-09-29 | | Release date: | 2011-10-19 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A two-in-one antibody against HER3 and EGFR has superior inhibitory activity compared with monospecific antibodies.
Cancer Cell, 20, 2011
|
|
3P11
 
 | |
8GW7
 
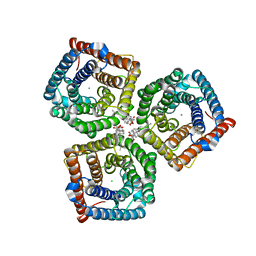 | | AtSLAC1 6D mutant in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein (Fragment) | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2022-09-16 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8GW6
 
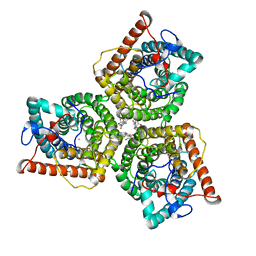 | | AtSLAC1 6D mutant in closed state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein (Fragment) | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2022-09-16 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8ISQ
 
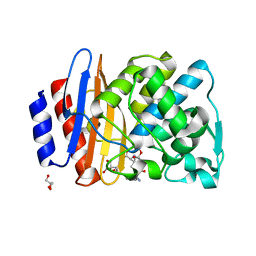 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISP
 
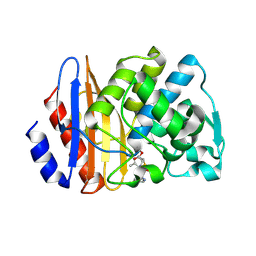 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cephalexin | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
