6H1F
 
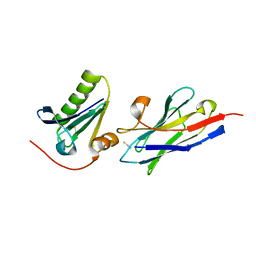 | | Structure of the nanobody-stabilized gelsolin D187N variant (second domain) | | Descriptor: | Gelsolin, THIOCYANATE ION, gelsolin nanobody, ... | | Authors: | Hassan, A, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2018-07-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nanobody interaction unveils structure, dynamics and proteotoxicity of the Finnish-type amyloidogenic gelsolin variant.
Biochim Biophys Acta Mol Basis Dis, 1865, 2019
|
|
6Q9Z
 
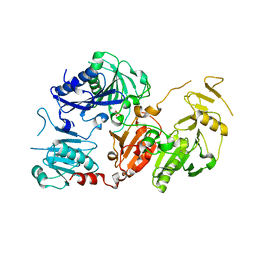 | | Crystal structure of the pathological G167R variant of calcium-free human gelsolin, | | Descriptor: | GLYCEROL, Gelsolin, SULFATE ION | | Authors: | Boni, F, Scalone, E, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6QBF
 
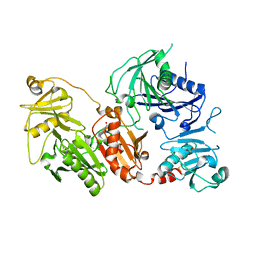 | | Crystal structure of the pathological D187N variant of calcium-free human gelsolin. | | Descriptor: | GLYCEROL, Gelsolin, SODIUM ION, ... | | Authors: | Scalone, E, Boni, F, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.499 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6Q9R
 
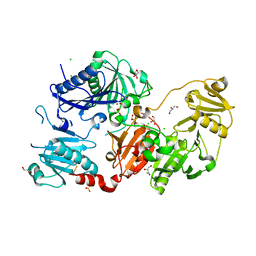 | | Crystal structure of the pathological N184K variant of calcium-free human gelsolin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Scalone, E, Boni, F, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6TCC
 
 | | Crystal structure of Salmo salar RidA-1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ricagno, S, Visentin, C, Di Pisa, F, Digiovanni, S, Oberti, L, Degani, G, Popolo, L, Bartorelli, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Two novel fish paralogs provide insights into the Rid family of imine deaminases active in pre-empting enamine/imine metabolic damage.
Sci Rep, 10, 2020
|
|
6TCD
 
 | | Crystal structure of Salmo salar RidA-2 | | Descriptor: | ACETATE ION, Ribonuclease UK114, SULFATE ION | | Authors: | Ricagno, S, Visentin, C, Di Pisa, F, Digiovanni, S, Oberti, L, Degani, G, Popolo, L, Bartorelli, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Two novel fish paralogs provide insights into the Rid family of imine deaminases active in pre-empting enamine/imine metabolic damage.
Sci Rep, 10, 2020
|
|
7ZS6
 
 | | Crystal structure of Apis mellifera RidA | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Reactive intermediate deaminase A, ... | | Authors: | Visentin, C, Rizzi, G, Ricagno, S. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Apis mellifera RidA, a novel member of the canonical YigF/YER057c/UK114 imine deiminase superfamily of enzymes pre-empting metabolic damage.
Biochem.Biophys.Res.Commun., 616, 2022
|
|
5FAF
 
 | | N184K pathological variant of gelsolin domain 2 (orthorhombic form) | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Boni, F, Milani, M, Ricagno, s, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
5O7G
 
 | |
5OLU
 
 | | The crystal structure of a highly thermostable carboxyl esterase from Bacillus coagulans in complex with glycerol | | Descriptor: | ACETATE ION, Alpha/beta hydrolase family protein, CHLORIDE ION, ... | | Authors: | Gourlay, L.J, Nakhnoukh, C, Bolognesi, M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A stereospecific carboxyl esterase from Bacillus coagulans hosting nonlipase activity within a lipase-like fold.
FEBS J., 285, 2018
|
|
5M6A
 
 | | Crystal structure of cardiotoxic Bence-Jones light chain dimer H9 | | Descriptor: | Bence-Jones light chain, GLYCEROL, PHOSPHATE ION | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5M76
 
 | | Crystal structure of cardiotoxic Bence-Jones light chain dimer H10 | | Descriptor: | BROMIDE ION, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-26 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5M6I
 
 | | Crystal structure of non-cardiotoxic Bence-Jones light chain dimer M8 | | Descriptor: | SODIUM ION, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
4RMS
 
 | |
6RQR
 
 | | Extended NHERF1 PDZ2 domain in complex with the PDZ-binding motif of CFTR | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1,Cystic fibrosis transmembrane conductance regulator | | Authors: | Martin, E.R, Ford, R.C, Robinson, R.C. | | Deposit date: | 2019-05-16 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In vivocrystals reveal critical features of the interaction between cystic fibrosis transmembrane conductance regulator (CFTR) and the PDZ2 domain of Na+/H+exchange cofactor NHERF1.
J.Biol.Chem., 295, 2020
|
|
5FAE
 
 | | N184K pathological variant of gelsolin domain 2 (trigonal form) | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boni, F, Milani, M, Ricagno, S, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
3V4Q
 
 | | Structure of R335W mutant of human Lamin | | Descriptor: | Prelamin-A/C | | Authors: | Bollati, M, Bolognesi, M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structures of the lamin A/C R335W and E347K mutants: Implications for dilated cardiolaminopathies.
Biochem.Biophys.Res.Commun., 418, 2012
|
|
3V4W
 
 | | Structure of E347K mutant of Lamin | | Descriptor: | Prelamin-A/C | | Authors: | Bollati, M, Bolognesi, M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structures of the lamin A/C R335W and E347K mutants: Implications for dilated cardiolaminopathies.
Biochem.Biophys.Res.Commun., 418, 2012
|
|
3NA4
 
 | | D53P beta-2 microglobulin mutant | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Beta-2-microglobulin | | Authors: | Azinas, S, Ricagno, S, Bolognesi, M. | | Deposit date: | 2010-06-01 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-strand perturbation and amyloid propensity in beta-2 microglobulin
Febs J., 278, 2011
|
|
3V5B
 
 | | Structure of Coil 2b of human lamin | | Descriptor: | Prelamin-A/C | | Authors: | Bollati, M, Bolognesi, M. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the lamin A/C R335W and E347K mutants: Implications for dilated cardiolaminopathies.
Biochem.Biophys.Res.Commun., 418, 2012
|
|
5MVG
 
 | | Crystal structure of non-amyloidogenic light chain dimer M7 | | Descriptor: | GLYCEROL, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-16 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5MTL
 
 | | Crystal structure of an amyloidogenic light chain | | Descriptor: | light chain dimer,IGL@ protein,IGL@ protein | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-10 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5MUD
 
 | | Crystal structure of an amyloidogenic light chain dimer H6 | | Descriptor: | light chain dimer,IGL@ protein | | Authors: | Oberti, L, Bacarizo, J, Maritan, M, Rognoni, P, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5O2Z
 
 | | Domain swap dimer of the G167R variant of gelsolin second domain | | Descriptor: | ACETATE ION, CALCIUM ION, CITRATE ANION, ... | | Authors: | Boni, F, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gelsolin pathogenic Gly167Arg mutation promotes domain-swap dimerization of the protein.
Hum. Mol. Genet., 27, 2018
|
|
5MUH
 
 | | Crystal structure of an amyloidogenic light chain dimer H7 | | Descriptor: | light chain dimer | | Authors: | Oberti, L, Rognoni, P, Russo, R, Maritan, M, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
