7DO4
 
 | | Crystal structure of CD97-CD55 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Niu, M, Song, G. | | Deposit date: | 2020-12-11 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for CD97 recognition of the decay-accelerating factor CD55 suggests mechanosensitive activation of adhesion GPCRs.
J.Biol.Chem., 296, 2021
|
|
7BVP
 
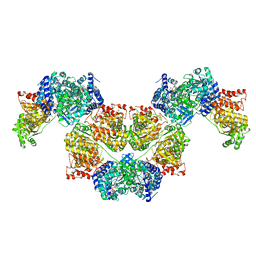 | | AdhE spirosome in extended conformation | | Descriptor: | Aldehyde-alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Kim, G.J, Song, J.J. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Aldehyde-alcohol dehydrogenase undergoes structural transition to form extended spirosomes for substrate channeling.
Commun Biol, 3, 2020
|
|
7EL7
 
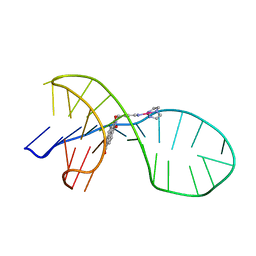 | | NMR solution structure of the 1:1 complex of a quadruplex-duplex hybrid MYT1L and a platinum(II) ligand L1Pt(dien) | | Descriptor: | G-quadruplex DNA MYT1L, Pt(diethylenetriamine)(2-(pyridin-4-ylmethyl)benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) | | Authors: | Liu, L.-Y, Liu, W, Mao, Z.-W. | | Deposit date: | 2021-04-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spatial Matching Selectivity and Solution Structure of Organic-Metal Hybrid to Quadruplex-Duplex Hybrid.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7FB3
 
 | |
7FB0
 
 | |
7FB4
 
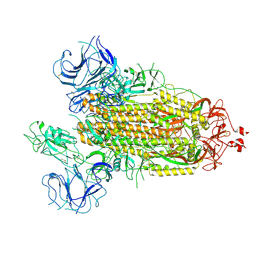 | |
7FB1
 
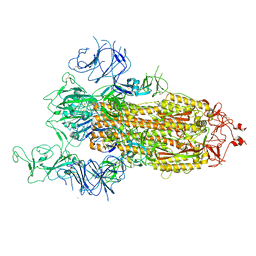 | |
8VHH
 
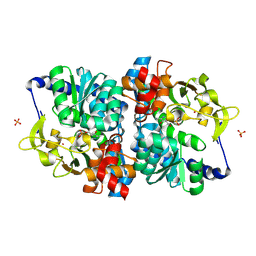 | | Engineered holo tryptophan synthase (Tm9D8*) derived from T. maritima TrpB | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Porter, N.J, Johnston, K.E, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2024-01-02 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A combinatorially complete epistatic fitness landscape in an enzyme active site.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7DEK
 
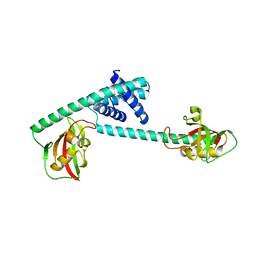 | | Pseudomonas aeruginosa FK506-binding protein PaFkbA | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Ynag, J, Huang, Q, Bao, R. | | Deposit date: | 2020-11-04 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization of PaFkbA: A periplasmic chaperone from Pseudomonas aeruginosa .
Comput Struct Biotechnol J, 19, 2021
|
|
2YPT
 
 | | Crystal structure of the human nuclear membrane zinc metalloprotease ZMPSTE24 mutant (E336A) in complex with a synthetic CSIM tetrapeptide from the C-terminus of prelamin A | | Descriptor: | CAAX PRENYL PROTEASE 1 HOMOLOG, PRELAMIN-A/C, ZINC ION | | Authors: | Pike, A.C.W, Dong, Y.Y, Quigley, A, Dong, L, Savitsky, P, Cooper, C.D.O, Chaikuad, A, Goubin, S, Shrestha, L, Li, Q, Mukhopadhyay, S, Yang, J, Xia, X, Shintre, C.A, Barr, A.J, Berridge, G, Chalk, R, Bray, J.E, von Delft, F, Bullock, A, Bountra, C, Arrowsmith, C.H, Edwards, A, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2012-11-01 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Structural Basis of Zmpste24-Dependent Laminopathies.
Science, 339, 2013
|
|
7WEX
 
 | |
6JKJ
 
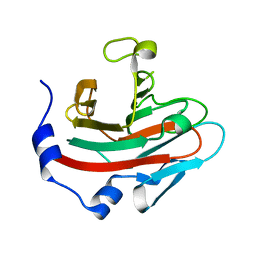 | | Crystal structure of human SPSB2 in the apo-state | | Descriptor: | SPRY domain-containing SOCS box protein 2 | | Authors: | Luo, Y, Kuang, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SPRY domain of human SPSB2 in the apo state.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7XDQ
 
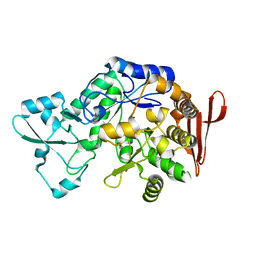 | | Crystal structure of a glucosylglycerol phosphorylase mutant from Marinobacter adhaerens | | Descriptor: | Glucosylglycerol phosphorylase, LITHIUM ION, beta-D-glucopyranose | | Authors: | Wei, H.L, Li, Q, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Protein Engineering of Glucosylglycerol Phosphorylase Facilitating Efficient and Highly Regio- and Stereoselective Glycosylation of Polyols in a Synthetic System.
Acs Catalysis, 2022
|
|
7XDR
 
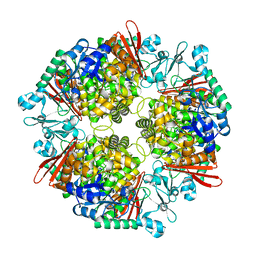 | | Crystal structure of a glucosylglycerol phosphorylase from Marinobacter adhaerens | | Descriptor: | Glucosylglycerol phosphorylase | | Authors: | Wei, H.L, Li, Q, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein Engineering of Glucosylglycerol Phosphorylase Facilitating Efficient and Highly Regio- and Stereoselective Glycosylation of Polyols in a Synthetic System.
Acs Catalysis, 2022
|
|
6K9V
 
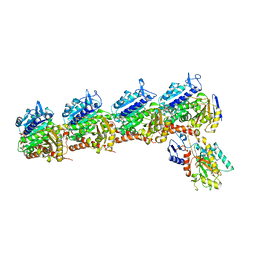 | | Crystal structure of tubulin in complex with inhibitor D64 | | Descriptor: | (5-methoxy-1H-indol-2-yl)-phenyl-methanone, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2019-06-18 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Structural insights into the design of indole derivatives as tubulin polymerization inhibitors.
Febs Lett., 594, 2020
|
|
5Z55
 
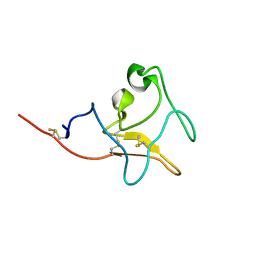 | |
6KOX
 
 | |
6KOW
 
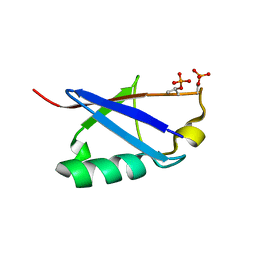 | |
7YGV
 
 | | Crystal structure of the Ca2+-bound EFhd1/Swiprosin-2 | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D1, GLYCEROL, ... | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Ynag, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
6KNZ
 
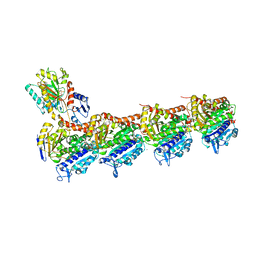 | | Crystal structure of T2R-TTL-KXO1 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[5-[4-(2-morpholin-4-ylethoxy)phenyl]pyridin-2-yl]-~{N}-(phenylmethyl)ethanamide, CALCIUM ION, ... | | Authors: | Chen, Q, Yu, Y. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.475 Å) | | Cite: | Reversible binding of the anticancer drug KXO1 (tirbanibulin) to the colchicine-binding site of beta-tubulin explains KXO1's low clinical toxicity.
J.Biol.Chem., 294, 2019
|
|
6KEY
 
 | |
6BLQ
 
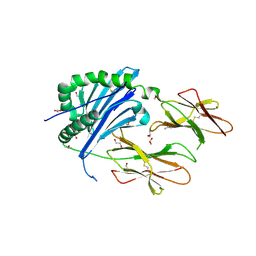 | | Crystal Structure of IAg7 in complex with insulin mimotope p8E9E | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2017-11-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-terminal modification of the insulin B:11-23 peptide creates superagonists in mouse and human type 1 diabetes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8HIJ
 
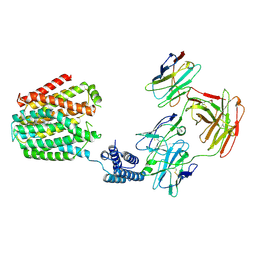 | | The 5-MTHF-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HIK
 
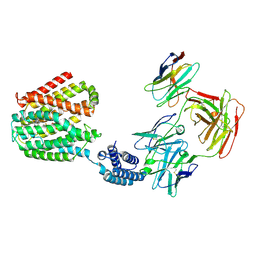 | | The TPP-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HII
 
 | | The BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | BRIL-SLC19A1 chimera, anti-BRIL Fab heavy chain, anti-BRIL Fab light chain, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
