8C3E
 
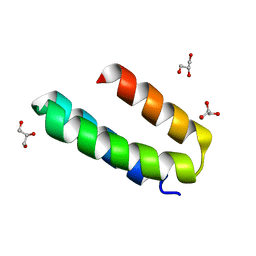 | | Engineered mini-protein LCB2 (blocking ligand of SARS-Cov-2 spike protein) | | Descriptor: | Engineered protein LCB2, GLYCEROL | | Authors: | Korban, S.A, Mikhailovskii, O.V, Luzik, D.A, Gurzhiy, V.V, Levkina, A.D, Kharkov, B.B, Skrynnikov, N.R. | | Deposit date: | 2022-12-23 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineered mini-protein LCB2 (blocking ligand of SARS-Cov-2 spike protein)
To Be Published
|
|
1UJZ
 
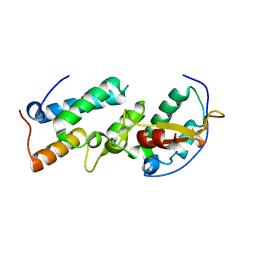 | | Crystal structure of the E7_C/Im7_C complex; a computationally designed interface between the colicin E7 DNase and the Im7 Immunity protein | | Descriptor: | Designed Colicin E7 DNase, Designed Colicin E7 immunity protein | | Authors: | Kortemme, T, Joachimiak, L.A, Bullock, A.N, Schuler, A.D, Stoddard, B.L, Baker, D. | | Deposit date: | 2003-08-13 | | Release date: | 2004-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computational redesign of protein-protein interaction specificity
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
1KER
 
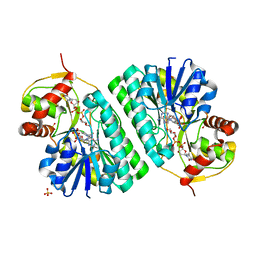 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with dTDP-D-glucose bound | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
1R8P
 
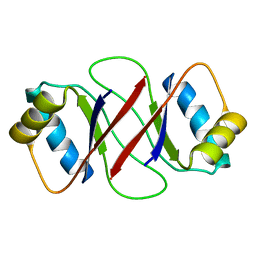 | | HPV-16 E2C solution structure | | Descriptor: | Regulatory protein E2 | | Authors: | Nadra, A.D, Eliseo, T, Cicero, D.O. | | Deposit date: | 2003-10-28 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HPV-16 E2 DNA binding domain, a transcriptional regulator with a dimeric beta-barrel fold
J.Biomol.NMR, 30, 2004
|
|
1R4P
 
 | | Shiga toxin type 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, FORMIC ACID, ... | | Authors: | Fraser, M.E, Fujinaga, M, Cherney, M.M, Melton-Celsa, A.R, Twiddy, E.M, O'Brien, A.D, James, M.N.G. | | Deposit date: | 2003-10-07 | | Release date: | 2004-05-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of Shiga Toxin Type 2 (Stx2) from Escherichia coli O157:H7.
J.Biol.Chem., 279, 2004
|
|
1IFU
 
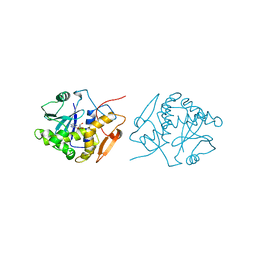 | | RICIN A-CHAIN (RECOMBINANT) COMPLEX WITH FORMYCIN | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, RICIN | | Authors: | Weston, S.A, Tucker, A.D, Thatcher, D.R, Derbyshire, D.J, Pauptit, R.A. | | Deposit date: | 1996-07-05 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of recombinant ricin A-chain at 1.8 A resolution.
J.Mol.Biol., 244, 1994
|
|
1RU9
 
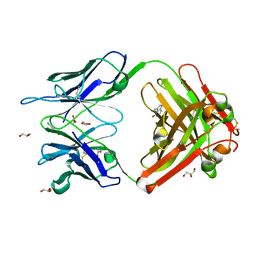 | | Crystal Structure (A) of u.v.-irradiated cationic cyclization antibody 4C6 Fab at pH 4.6 with a data set collected in-house. | | Descriptor: | ACETATE ION, BENZOIC ACID, GLYCEROL, ... | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
1RUK
 
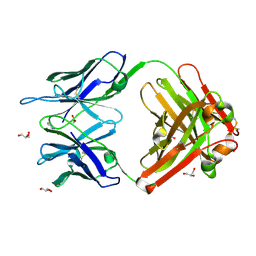 | | Crystal structure (C) of native cationic cyclization antibody 4C6 fab at pH 4.6 with a data set collected at SSRL beamline 9-1 | | Descriptor: | ACETATE ION, BENZOIC ACID, GLYCEROL, ... | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
7SIM
 
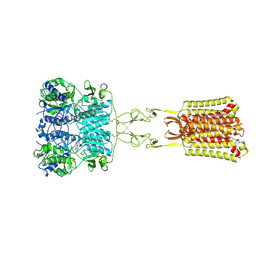 | | Structure of positive allosteric modulator-free active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SIN
 
 | | Structure of negative allosteric modulator-bound inactive human calcium-sensing receptor | | Descriptor: | 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, Isoform 1 of Extracellular calcium-sensing receptor | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8E9N
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIY in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9M
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT bound to maleic acid at 278 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9R
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFCS in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9L
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9O
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIY bound to maleic acid at 278 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9T
 
 | | Crystal structure of wild-type E. coli aspartate aminotransferase in the ligand-free form at 303 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9S
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFCS bound to maleic acid at 278 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9K
 
 | | Crystal structure of wild-type E. coli aspartate aminotransferase bound to maleate at 278 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
1R4Q
 
 | | Shiga toxin | | Descriptor: | SHT cytotoxin A subunit, Shigella toxin chain B | | Authors: | Fraser, M.E, Fujinaga, M, Cherney, M.M, Melton-Celsa, A.R, Twiddy, E.M, O'Brien, A.D, James, M.N.G. | | Deposit date: | 2003-10-07 | | Release date: | 2004-05-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Shiga Toxin Type 2 (Stx2) from Escherichia coli O157:H7.
J.Biol.Chem., 279, 2004
|
|
8E9Q
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX bound to maleic acid at 278 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9V
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT in the ligand-free form at 303 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9J
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9P
 
 | | Crystal structure of wild-type E. coli aspartate aminotransferase in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9C
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant AIFS in the ligand-free form at 100 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9D
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant AIFS bound to maleic acid at 100 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
