8XH6
 
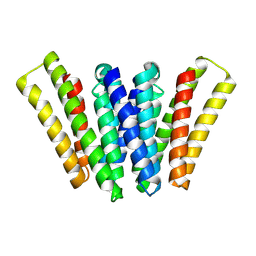 | | Structure of EBV LMP1 dimer | | Descriptor: | Latent membrane protein 1 | | Authors: | Gao, P, Huang, J.F. | | Deposit date: | 2023-12-17 | | Release date: | 2024-06-26 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Assembly and activation of EBV latent membrane protein 1.
Cell, 187, 2024
|
|
8WH4
 
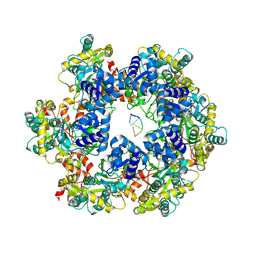 | | MPOX E5 hexamer ssDNA bound apo conformation | | Descriptor: | DNA (5'-D(P*CP*CP*CP*C)-3'), Uncoating factor OPG117 | | Authors: | Zhang, Z, Dong, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer.
Sci Adv, 10, 2024
|
|
8WH3
 
 | | MPOX E5 hexamer apo form | | Descriptor: | Uncoating factor OPG117 | | Authors: | Zhang, Z, Dong, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer.
Sci Adv, 10, 2024
|
|
8WH6
 
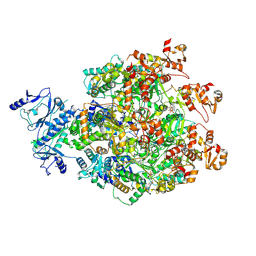 | |
8WGZ
 
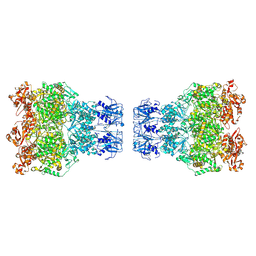 | | MPOX E5 double hexamer ssDNA bound conformation | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*C)-3'), Uncoating factor OPG117 | | Authors: | Zhang, Z, Dong, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer.
Sci Adv, 10, 2024
|
|
8WGY
 
 | |
8WH0
 
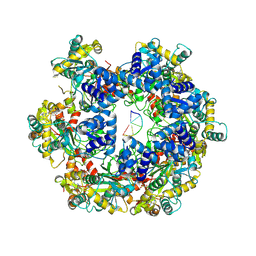 | | MPOX E5 hexamer ssDNA and AMP-PNP bound conformation | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*C)-3'), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhang, Z, Dong, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer.
Sci Adv, 10, 2024
|
|
6IIH
 
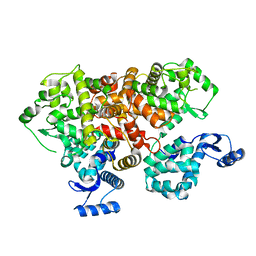 | | crystal structure of mitochondrial calcium uptake 2(MICU2) | | Descriptor: | CALCIUM ION, Endolysin,Calcium uptake protein 2, mitochondrial | | Authors: | Shen, Q, Wu, W, Zheng, J, Jia, Z. | | Deposit date: | 2018-10-06 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | The crystal structure of MICU2 provides insight into Ca2+binding and MICU1-MICU2 heterodimer formation.
Embo Rep., 20, 2019
|
|
6K2O
 
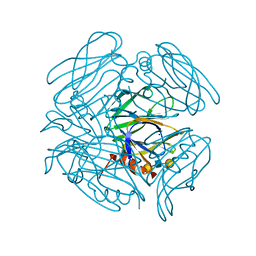 | | Structural basis of glycan recognition in globally predominant human P[8] rotavirus | | Descriptor: | Outer capsid protein VP4, SODIUM ION, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose | | Authors: | Duan, Z, Sun, X. | | Deposit date: | 2019-05-15 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Structural Basis of Glycan Recognition in Globally Predominant Human P[8] Rotavirus.
Virol Sin, 35, 2020
|
|
6KG6
 
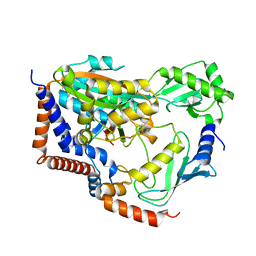 | | Crystal structure of MavC/UBE2N-Ub complex | | Descriptor: | MavC, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 N | | Authors: | Wang, Y, Huang, Y, Chang, M, Feng, Y. | | Deposit date: | 2019-07-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
6K2N
 
 | |
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | Descriptor: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
7TMC
 
 | |
7V47
 
 | |
7V48
 
 | |
7V49
 
 | |
7VCP
 
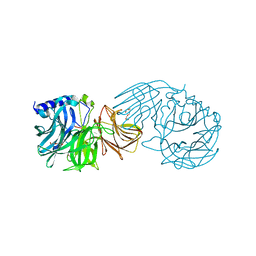 | | Frischella perrara beta-fructofuranosidase in complex with fructose | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Sucrose-6-phosphate hydrolase, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic and structural characterization of beta-fructofuranosidase from the honeybee gut bacterium Frischella perrara.
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7VCO
 
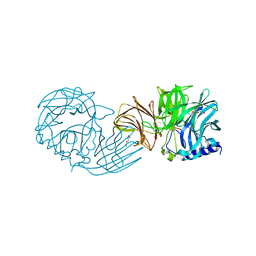 | | Frischella perrara beta-fructofuranosidase | | Descriptor: | DI(HYDROXYETHYL)ETHER, Sucrose-6-phosphate hydrolase, TRIETHYLENE GLYCOL | | Authors: | Tonozuka, T. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzymatic and structural characterization of beta-fructofuranosidase from the honeybee gut bacterium Frischella perrara.
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7UN5
 
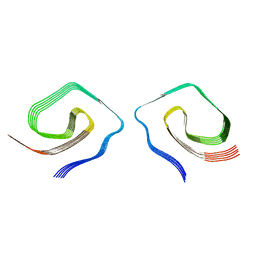 | |
7UMQ
 
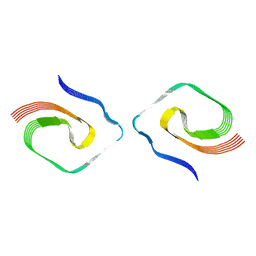 | |
7DRK
 
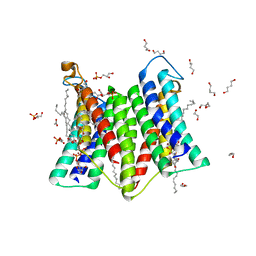 | | Crystal structure of phosphatidylglycerol phosphate synthase in complex with cytidine diphosphate-diacylglycerol | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,4-BUTANEDIOL, 5'-O-[(R)-{[(S)-{(2R)-2,3-bis[(9E)-octadec-9-enoyloxy]propoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, ... | | Authors: | Yang, B.W, Liu, Z.F. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The phosphatidylglycerol phosphate synthase PgsA utilizes a trifurcated amphipathic cavity for catalysis at the membrane-cytosol interface.
Curr Res Struct Biol, 3, 2021
|
|
7DRJ
 
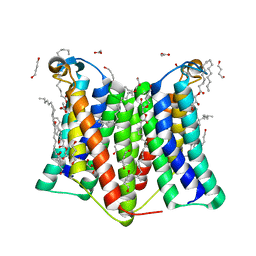 | | Crystal structure of phosphatidylglycerol phosphate synthase in complex with phosphatidylglycerol phosphate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,4-BUTANEDIOL, ACETIC ACID, ... | | Authors: | Yang, B.W, Liu, Z.F. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The phosphatidylglycerol phosphate synthase PgsA utilizes a trifurcated amphipathic cavity for catalysis at the membrane-cytosol interface.
Curr Res Struct Biol, 3, 2021
|
|
7EG0
 
 | | Cryo-EM structure of anagrelide-induced PDE3A-SLFN12 complex | | Descriptor: | 6,7-bis(chloranyl)-3,5-dihydro-1H-imidazo[2,1-b]quinazolin-2-one, MAGNESIUM ION, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-23 | | Release date: | 2021-09-29 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7EG4
 
 | | Cryo-EM structure of nauclefine-induced PDE3A-SLFN12 complex | | Descriptor: | MAGNESIUM ION, Parvine, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7E0F
 
 | |
