4BVF
 
 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH THIOALKYLIMIDATE FORMED FROM THIO-ACETYL-LYSINE ACS2-PEPTIDE CRYSTALLIZED IN PRESENCE OF THE INHIBITOR EX-527 | | Descriptor: | 1,2-ETHANEDIOL, ACETYL-COENZYME A SYNTHETASE 2-LIKE, MITOCHONDRIAL, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6U32
 
 | |
6U2M
 
 | | Crystal structure of a HaloTag-based calcium indicator, HaloCaMP V2, bound to JF635 | | Descriptor: | (1E,3S)-1-{10-[2-carboxy-5-({2-[2-(hexyloxy)ethoxy]ethyl}carbamoyl)phenyl]-7-(3-fluoroazetidin-1-yl)-5,5-dimethyldibenz o[b,e]silin-3(5H)-ylidene}-3-fluoroazetidin-1-ium, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Deo, C, Schreiter, E.R. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The HaloTag as a general scaffold for far-red tunable chemigenetic indicators.
Nat.Chem.Biol., 17, 2021
|
|
5OAI
 
 | | Structure of MDM2 with low molecular weight inhibitor | | Descriptor: | 3-[(1~{R})-2-(~{tert}-butylamino)-1-[methanoyl-[[3,4,5-tris(fluoranyl)phenyl]methyl]amino]-2-oxidanylidene-ethyl]-6-chloranyl-1~{H}-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Neochoritis, C.G, Grudnik, P, Dubin, G, Domling, A, Holak, T.A. | | Deposit date: | 2017-06-22 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A fluorinated indole-based MDM2 antagonist selectively inhibits the growth of p53wtosteosarcoma cells.
Febs J., 286, 2019
|
|
6OLU
 
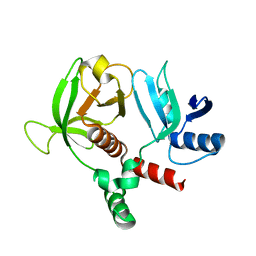 | | RIAM RA-PH core structure in the P212121 space group | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | | Authors: | Wu, J. | | Deposit date: | 2019-04-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphorylation of RIAM by src promotes integrin activation by unmasking the PH domain of RIAM.
Structure, 29, 2021
|
|
7BK4
 
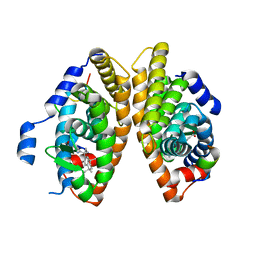 | | Crystal structure of RXRalpha ligand binding domain in complex with a fragment of the TIF2 coactivator | | Descriptor: | 6-[1-(3,5,5,8,8-PENTAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)CYCLOPROPYL]PYRIDINE-3-CARBOXYLIC ACID, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | le Maire, A, Bourguet, W. | | Deposit date: | 2021-01-15 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Interaction of the Intrinsically Disordered Co-activator TIF2 with Retinoic Acid Receptor Heterodimer (RXR/RAR).
J.Mol.Biol., 433, 2021
|
|
6O6H
 
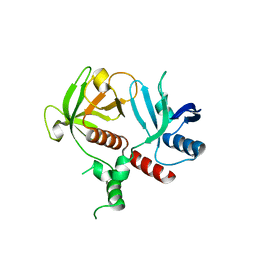 | | RIAM cc-RA-PH structure in the P21212 space group | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | | Authors: | Wu, J. | | Deposit date: | 2019-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphorylation of RIAM by Src Promotes Integrin Activation by Unmasking the PH Domain of RIAM.
Structure, 2020
|
|
4YTC
 
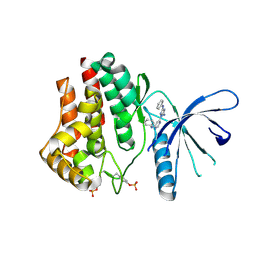 | |
6QKL
 
 | |
4YTI
 
 | |
3FMA
 
 | |
6BQK
 
 | |
6BQJ
 
 | |
3ZEV
 
 | | Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | GLYCINE, NEUROTENSIN, NEUROTENSIN RECEPTOR 1 TM86V | | Authors: | Egloff, P, Hillenbrand, M, Schlinkmann, K.M, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-29 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3GMD
 
 | | Structure-based design of 7-Azaindole-pyrrolidines as inhibitors of 11beta-Hydroxysteroid-Dehydrogenase type I | | Descriptor: | 2-methyl-3-{(3S)-1-[(1-pyridin-2-ylcyclopropyl)carbonyl]pyrrolidin-3-yl}-1H-pyrrolo[2,3-b]pyridine, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Valeur, E, Lepifre, F, Roche, D, Christmann-Franck, S, Hillertz, P, Musil, D. | | Deposit date: | 2009-03-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-based design of 7-azaindole-pyrrolidine amides as inhibitors of 11 beta-hydroxysteroid dehydrogenase type I.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6YG8
 
 | |
6YBB
 
 | | Crystal structure of a native BcsE (217-523) - BcsR-BcsQ (R156E mutant) complex with c-di-GMP and ATP bound | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ADENOSINE-5'-TRIPHOSPHATE, Bacterial cellulose secretion regulator BcsE, ... | | Authors: | Abidi, W, Zouhir, S, Roche, S, Krasteva, P.V. | | Deposit date: | 2020-03-16 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Architecture and regulation of an enterobacterial cellulose secretion system.
Sci Adv, 7, 2021
|
|
6YBU
 
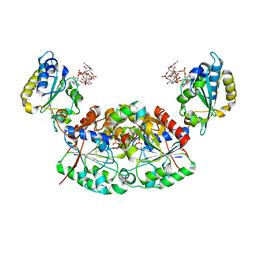 | | Crystal structure of a native BcsE (349-523) RQ complex with c-di-GMP and ATP bound | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ADENOSINE-5'-TRIPHOSPHATE, Bacterial cellulose secretion regulator BcsE, ... | | Authors: | Abidi, W, Zouhir, S, Roche, S, Krasteva, P.V. | | Deposit date: | 2020-03-17 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Architecture and regulation of an enterobacterial cellulose secretion system.
Sci Adv, 7, 2021
|
|
6Z72
 
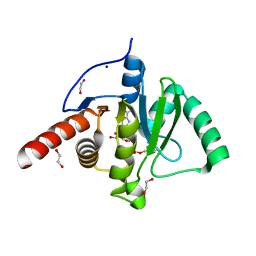 | | SARS-CoV-2 Macrodomain in complex with ADP-HPM | | Descriptor: | 1,2-ETHANEDIOL, Adenosine Diphosphate (Hydroxymethyl)pyrrolidine monoalcohol, D-MALATE, ... | | Authors: | Zorzini, V, Rack, J, Ahel, I. | | Deposit date: | 2020-05-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential.
Open Biology, 10, 2020
|
|
6Z5T
 
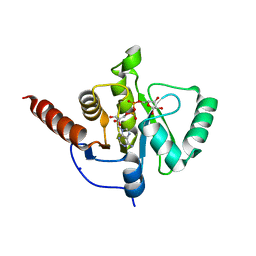 | | SARS-CoV-2 Macrodomain in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Replicase polyprotein 1ab, SODIUM ION | | Authors: | Zorzini, V, Rack, J, Ahel, I. | | Deposit date: | 2020-05-27 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential.
Open Biology, 10, 2020
|
|
4BUO
 
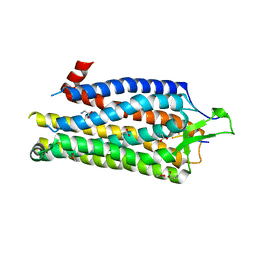 | | High Resolution Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | GLYCINE, NEUROTENSIN RECEPTOR TYPE 1, NEUROTENSIN/NEUROMEDIN N | | Authors: | Egloff, P, Hillenbrand, M, Schlinkmann, K.M, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2013-06-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BWB
 
 | | Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | NEUROTENSIN, NEUROTENSIN RECEPTOR TYPE 1 | | Authors: | Egloff, P, Hillenbrand, M, Scott, D.J, Schlinkmann, K.M, Heine, P, Balada, S, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2013-07-01 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4C4V
 
 | |
4C47
 
 | |
8QOD
 
 | | CRYSTAL STRUCTURE OF FVIIA IN COMPLEX WITH A BENZAMIDINE-BASED INHIBITOR | | Descriptor: | 3-[2-bromanyl-4-[(1~{S})-1-[[3-[(4-carbamimidoylphenyl)amino]-3-oxidanylidene-propanoyl]amino]ethyl]phenoxy]benzoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2023-09-28 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Crystallography and molecular simulations capture S1 pocket
collapse in allosteric regulation of factor VIIa and other serine
proteases.
To be published
|
|
