5XLR
 
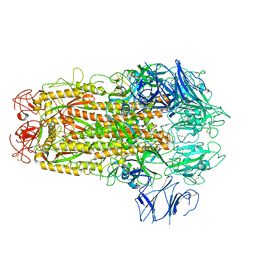 | | Structure of SARS-CoV spike glycoprotein | | Descriptor: | Spike glycoprotein | | Authors: | Gui, M, Song, W, Xiang, Y, Wang, X. | | Deposit date: | 2017-05-11 | | Release date: | 2017-06-07 | | Last modified: | 2019-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding.
Cell Res., 27, 2017
|
|
7BCY
 
 | |
7BED
 
 | |
7C6Q
 
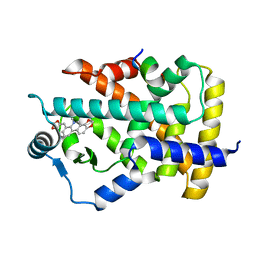 | | Novel natural PPARalpha agonist with a unique binding mode | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Peroxisome proliferator-activated receptor alpha | | Authors: | Tian, S.Y, Wang, R, Zheng, W.L, Li, Y. | | Deposit date: | 2020-05-22 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural Basis for PPARs Activation by The Dual PPAR alpha / gamma Agonist Sanguinarine: A Unique Mode of Ligand Recognition.
Molecules, 26, 2021
|
|
7FH0
 
 | |
4RAP
 
 | | Crystal structure of bacterial iron-containing dodecameric glycosyltransferase TibC from enterotoxigenic E.coli H10407 | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Glycosyltransferase TibC | | Authors: | Yao, Q, Lu, Q, Xu, Y, Shao, F. | | Deposit date: | 2014-09-10 | | Release date: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | A structural mechanism for bacterial autotransporter glycosylation by a dodecameric heptosyltransferase family.
Elife, 3, 2014
|
|
6IVN
 
 | | Crystal structure of a membrane protein G264A | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IVL
 
 | | Crystal structure of a membrane protein L259A | | Descriptor: | ACETIC ACID, CHLORIDE ION, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IOM
 
 | | Crystal structure of human C4.4A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ly6/PLAUR domain-containing protein 3 | | Authors: | Huang, M.D, Jiang, Y.B, Yuan, C, Lin, L. | | Deposit date: | 2018-10-30 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Crystal Structures of Human C4.4A Reveal the Unique Association of Ly6/uPAR/alpha-neurotoxin Domain
Int J Biol Sci, 16, 2020
|
|
6IVJ
 
 | | Crystal structure of a membrane protein G18A | | Descriptor: | ACETIC ACID, CHLORIDE ION, Ibestrophin, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6NJM
 
 | | Architecture and subunit arrangement of native AMPA receptors | | Descriptor: | 15F1 Fab heavy chain, 15F1 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gouaux, E, Zhao, Y. | | Deposit date: | 2019-01-03 | | Release date: | 2019-04-24 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM.
Science, 364, 2019
|
|
6NJL
 
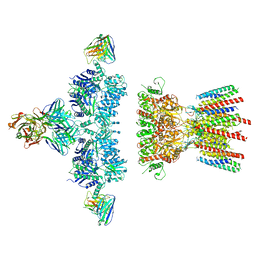 | |
6NJN
 
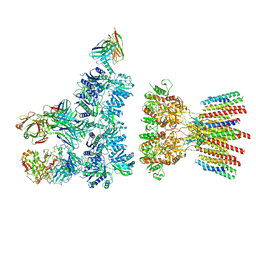 | |
6OGJ
 
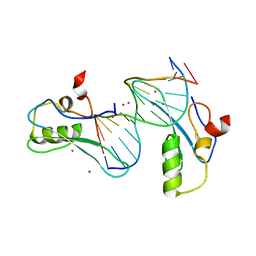 | | MeCP2 MBD in complex with DNA | | Descriptor: | DNA (5'-D(*CP*GP*GP*AP*GP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*CP*AP*CP*TP*CP*CP*G)-3'), Methyl-CpG-binding protein 2, ... | | Authors: | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
7WIN
 
 | |
7Y09
 
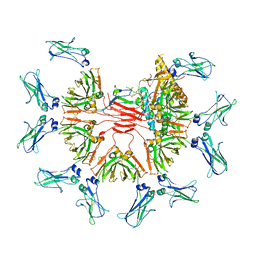 | | Cryo-EM structure of human IgM-Fc in complex with the J chain and the DBL domain of DBLMSP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Shen, H, Ji, C, Xiao, J. | | Deposit date: | 2022-06-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Plasmodium falciparum has evolved multiple mechanisms to hijack human immunoglobulin M.
Nat Commun, 14, 2023
|
|
7YG2
 
 | | Cryo-EM structure of human IgM-Fc in complex with the J chain and the DBL domain of DBLMSP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DBLMSP2, ... | | Authors: | Shen, H, Ji, C, Xiao, J. | | Deposit date: | 2022-07-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Plasmodium falciparum has evolved multiple mechanisms to hijack human immunoglobulin M.
Nat Commun, 14, 2023
|
|
7Y0J
 
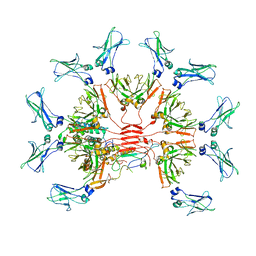 | |
7Y0H
 
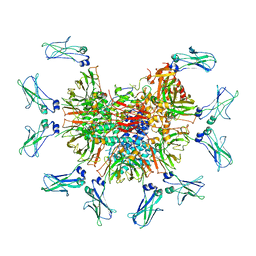 | |
7Y4H
 
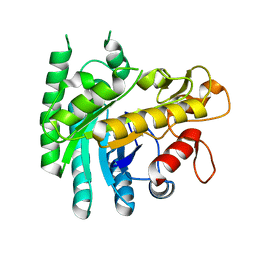 | |
7FEO
 
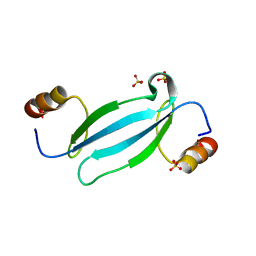 | | Crystal structure of AtMBD5 MBD domain | | Descriptor: | Methyl-CpG-binding domain-containing protein 5, SULFATE ION | | Authors: | Zhou, M.Q, Wu, Z.B, Liu, K, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Family-wide Characterization of Methylated DNA Binding Ability of Arabidopsis MBDs.
J.Mol.Biol., 434, 2022
|
|
7FEF
 
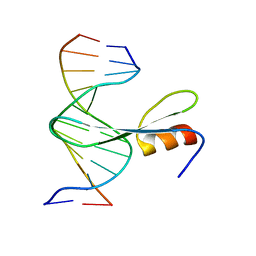 | | Crystal structure of AtMBD6 with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain-containing protein 6 | | Authors: | Wu, Z.B, Liu, K, Min, J.R. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Family-wide Characterization of Methylated DNA Binding Ability of Arabidopsis MBDs.
J.Mol.Biol., 434, 2022
|
|
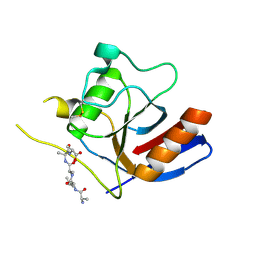
7F2D
 
 | |
7F2I
 
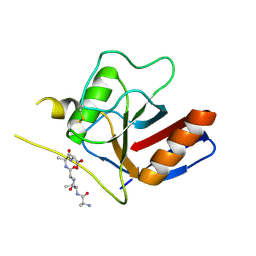 | |
3O98
 
 | | Glutathionylspermidine synthetase/amidase C59A complex with ADP and Gsp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, GLUTATHIONYLSPERMIDINE, ... | | Authors: | Pai, C.H, Lin, C.H, Wang, A.H.-J. | | Deposit date: | 2010-08-04 | | Release date: | 2011-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of Escherichia coli glutathionylspermidine amidase belonging to the family of cysteine; histidine-dependent amidohydrolases/peptidases
Protein Sci., 20, 2011
|
|
