4DKO
 
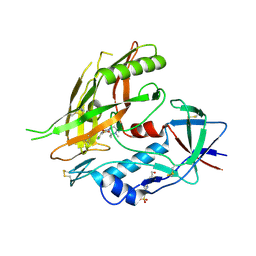 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with TS-II-224 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core, ... | | Authors: | Kwon, Y.D, LaLonde, J.M, Jones, D.M, Sun, A.W, Courter, J.R, Soeta, T, Kobayashi, T, Princiotto, A.M, Wu, X, Mascola, J, Schon, A, Freire, E, Sodroski, J, Madani, N, Smith III, A.B, Kwong, P.D. | | Deposit date: | 2012-02-03 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of Dual Hotspot Small-Molecule HIV-1 Entry Inhibitors.
J.Med.Chem., 55, 2012
|
|
3BDP
 
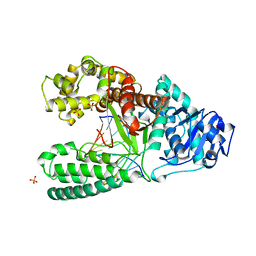 | | DNA POLYMERASE I/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*GP*CP*AP*TP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*GP*CP*2DT)-3'), PROTEIN (DNA POLYMERASE I), ... | | Authors: | Kiefer, J.R, Mao, C, Beese, L.S. | | Deposit date: | 1997-11-17 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal.
Nature, 391, 1998
|
|
6CO2
 
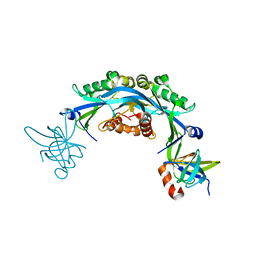 | | Structure of an engineered protein (NUDT16TI) in complex with 53BP1 Tudor domains | | Descriptor: | NUDT16-Tudor-interacting (NUDT16TI), TP53-binding protein 1 | | Authors: | Botuyan, M.V, Thompson, J.R, Cui, G, Mer, G. | | Deposit date: | 2018-03-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4R28
 
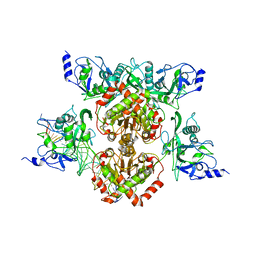 | |
2BDP
 
 | | CRYSTAL STRUCTURE OF BACILLUS DNA POLYMERASE I FRAGMENT COMPLEXED TO 9 BASE PAIRS OF DUPLEX DNA | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*AP*GP*CP*AP*TP*CP*AP*TP*GP*C)-3'), MAGNESIUM ION, ... | | Authors: | Kiefer, J.R, Mao, C, Beese, L.S. | | Deposit date: | 1997-11-17 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal.
Nature, 391, 1998
|
|
4Y7B
 
 | | Factor Xa complex with GTC000441 | | Descriptor: | 6-chloro-N-{(3S)-1-[(2S)-1-{(1R,5S)-7-[2-(methylamino)ethyl]-3,7-diazabicyclo[3.3.1]non-3-yl}-1-oxopropan-2-yl]-2-oxopy rrolidin-3-yl}naphthalene-2-sulfonamide, CALCIUM ION, Coagulation factor X | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Factor Xa complex with GTC000441
to be published
|
|
2FN2
 
 | | SOLUTION NMR STRUCTURE OF THE GLYCOSYLATED SECOND TYPE TWO MODULE OF FIBRONECTIN, 20 STRUCTURES | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FIBRONECTIN | | Authors: | Sticht, H, Pickford, A.R, Potts, J.R, Campbell, I.D. | | Deposit date: | 1997-08-06 | | Release date: | 1998-09-16 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the glycosylated second type 2 module of fibronectin.
J.Mol.Biol., 276, 1998
|
|
2BDM
 
 | | Structure of Cytochrome P450 2B4 with Bound Bifonazole | | Descriptor: | 1-[PHENYL-(4-PHENYLPHENYL)-METHYL]IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Zhao, Y, White, M.A, Muralidhara, B.K, Sun, L, Halpert, J.R, Stout, C.D. | | Deposit date: | 2005-10-20 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction.
J.Biol.Chem., 281, 2006
|
|
4UW8
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber with its intra-molecular chaperone domain | | Descriptor: | CITRATE ANION, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
4UW7
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber without its intra-molecular chaperone domain | | Descriptor: | GLYCEROL, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
4Y71
 
 | | Factor Xa complex with GTC000398 | | Descriptor: | 6-chloro-N-{(3S)-1-[(2S)-1-(4-methyl-5-oxo-1,4-diazepan-1-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}naphthalene-2-sulf onamide, CALCIUM ION, Coagulation factor X | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Factor Xa complex with GTC000398
To be Published
|
|
4Y7A
 
 | | Factor Xa complex with GTC000422 | | Descriptor: | CALCIUM ION, Coagulation factor X, MAGNESIUM ION, ... | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Factor Xa complex with GTC000422
to be published
|
|
5CNA
 
 | | REFINED STRUCTURE OF CONCANAVALIN A COMPLEXED WITH ALPHA-METHYL-D-MANNOPYRANOSIDE AT 2.0 ANGSTROMS RESOLUTION AND COMPARISON WITH THE SACCHARIDE-FREE STRUCTURE | | Descriptor: | CALCIUM ION, CHLORIDE ION, CONCANAVALIN A, ... | | Authors: | Naismith, J.H, Emmerich, C, Habash, J, Harrop, S.J, Helliwell, J.R, Hunter, W.N, Raftery, J, Kalb(Gilboa), A.J, Yariv, J. | | Deposit date: | 1994-02-11 | | Release date: | 1994-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structure of concanavalin A complexed with methyl alpha-D-mannopyranoside at 2.0 A resolution and comparison with the saccharide-free structure.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
2BLM
 
 | |
2Y6P
 
 | | Evidence for a Two-Metal-Ion-Mechanism in the Kdo- Cytidylyltransferase KdsB | | Descriptor: | 3-DEOXY-MANNO-OCTULOSONATE CYTIDYLYLTRANSFERASE, BETA-MERCAPTOETHANOL, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Schmidt, H, Mesters, J.R, Mamat, U, Hilgenfeld, R. | | Deposit date: | 2011-01-25 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evidence for a Two-Metal-Ion Mechanism in the Cytidyltransferase Kdsb, an Enzyme Involved in Lipopolysaccharide Biosynthesis.
Plos One, 6, 2011
|
|
2Y7X
 
 | | The discovery of potent and long-acting oral factor Xa inhibitors with tetrahydroisoquinoline and benzazepine P4 motifs | | Descriptor: | 6-CHLORO-N-[(3S)-1-(5-FLUORO-1,2,3,4-TETRAHYDROISOQUINOLIN-6-YL)-2-OXO-PYRROLIDIN-3-YL]NAPHTHALENE-2-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Watson, N.S, Adams, C, Belton, D, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Harling, J.D, Irving, W.R, Irvine, S, Kleanthous, S, McLay, I.M, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Discovery of Potent and Long-Acting Oral Factor Xa Inhibitors with Tetrahydroisoquinoline and Benzazepine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2YZ4
 
 | | The neutron structure of concanavalin A at 2.2 Angstroms | | Descriptor: | CALCIUM ION, Concanavalin A, MANGANESE (II) ION | | Authors: | Ahmed, H.U, Blakeley, M.P, Cianci, M, Hubbard, J.A, Helliwell, J.R. | | Deposit date: | 2007-05-02 | | Release date: | 2008-02-05 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | The determination of protonation states in proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2YPN
 
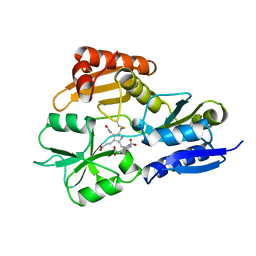 | | Hydroxymethylbilane synthase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, PROTEIN (HYDROXYMETHYLBILANE SYNTHASE) | | Authors: | Nieh, Y.P, Raftery, J, Weisgerber, S, Habash, J, Schotte, F, Ursby, T, Wulff, M, Haedener, A, Campbell, J.W, Hao, Q, Helliwell, J.R. | | Deposit date: | 1999-01-11 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Accurate and highly complete synchrotron protein crystal Laue diffraction data using the ESRF CCD and the Daresbury Laue software
J.Synchrotron Radiat., 6, 1999
|
|
2W9H
 
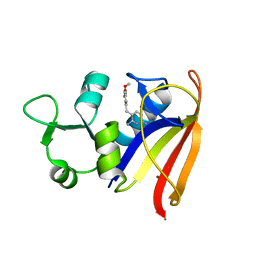 | |
6GNV
 
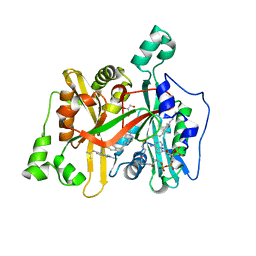 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and a isopropyl methyl indole aryl sulphonamide ligand | | Descriptor: | GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, ... | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6GQI
 
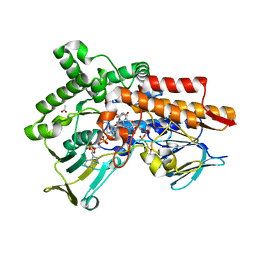 | |
2W9G
 
 | | Wild-type Staphylococcus aureus DHFR in complex with NADPH and trimethoprim | | Descriptor: | DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIMETHOPRIM | | Authors: | Soutter, H.H, Miller, J.R. | | Deposit date: | 2009-01-23 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Comparison of Chromosomal and Exogenous Dihydrofolate Reductase from Staphylococcus Aureus in Complex with the Potent Inhibitor Trimethoprim.
Proteins, 76, 2009
|
|
6GNH
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and an Azepanyl Phenyl Benzylsulphonamide Ligand | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, methyl 4-(azepan-1-yl)-3-[(4-methoxyphenyl)sulfonylamino]benzoate | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6GJQ
 
 | | human NBD1 of CFTR in complex with nanobody T27 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, Nanobody T27 | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-16 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
6GJT
 
 | |
