5CY9
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-30 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CRZ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)-4-fluorobenzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
8TBF
 
 | | Tricomplex of RMC-7977, KRAS WT, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 629, 2024
|
|
5AZF
 
 | | Crystal structure of LGG-1 complexed with a WEEL peptide | | Descriptor: | CADMIUM ION, Protein lgg-1, SULFATE ION, ... | | Authors: | Watanabe, Y, Noda, N.N. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
5AZG
 
 | | Crystal structure of LGG-1 complexed with a UNC-51 peptide | | Descriptor: | CADMIUM ION, Protein lgg-1, Serine/threonine-protein kinase unc-51 | | Authors: | Watanabe, Y, Fujioka, Y, Noda, N.N. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
9AX6
 
 | | Tricomplex of RMC-6236, KRAS G12D, and CypA | | Descriptor: | (1R,2S)-N-[(1P,7S,9S,13R,20M)-21-ethyl-20-{2-[(1R)-1-methoxyethyl]-5-(4-methylpiperazin-1-yl)pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-2-methylcyclopropane-1-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-03-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Translational and Therapeutic Evaluation of RAS-GTP Inhibition by RMC-6236 in RAS-Driven Cancers.
Cancer Discov, 14, 2024
|
|
5AZH
 
 | | Crystal structure of LGG-2 fused with an EEEWEEL peptide | | Descriptor: | EEEWEEL peptide,Protein lgg-2, MAGNESIUM ION | | Authors: | Watanabe, Y, Fujioka, Y, Noda, N.N. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
4M57
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 from maize | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10 | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
6EI3
 
 | | Crystal structure of auto inhibited POT family peptide transporter | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Proton-dependent oligopeptide transporter family protein | | Authors: | Newstead, S, Brinth, A, Vogeley, L, Caffrey, M. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Proton movement and coupling in the POT family of peptide transporters.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3CP1
 
 | |
2TIO
 
 | | LOW PACKING DENSITY FORM OF BOVINE BETA-TRYPSIN IN CYCLOHEXANE | | Descriptor: | BENZAMIDINE, CALCIUM ION, HEXANE, ... | | Authors: | Huang, Q, Zhu, G, Tang, Q. | | Deposit date: | 1998-09-23 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray studies on two forms of bovine beta-trypsin crystals in neat cyclohexane.
Biochim.Biophys.Acta, 1429, 1998
|
|
4M59
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 in complex with an 18-nt psaJ RNA element | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10, PHOSPHATE ION, psaJ RNA | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
8FE8
 
 | | Crystal Structure of HIV-1 RT in Complex with the non-nucleoside inhibitor 18b1 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}pyrimidin-2-yl)amino]-2-[4-(methanesulfonyl)piperazin-1-yl]benzonitrile, Reverse transcriptase p51, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of diarylpyrimidine derivatives bearing piperazine sulfonyl as potent HIV-1 nonnucleoside reverse transcriptase inhibitors.
Commun Chem, 6, 2023
|
|
3CYO
 
 | |
6JKM
 
 | | Crystal structure of BubR1 kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
6JKK
 
 | | Crystal structure of BubR1 kinase domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Mitotic checkpoint control protein kinase BUB1 | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
5CQR
 
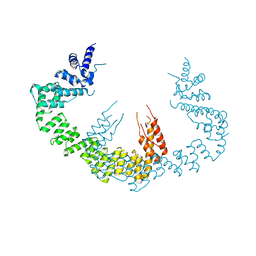 | | Dimerization of Elp1 is essential for Elongator complex assembly | | Descriptor: | Elongator complex protein 1 | | Authors: | Lin, Z, Xu, H, Li, F, Diao, W, Long, J, Shen, Y. | | Deposit date: | 2015-07-22 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.015 Å) | | Cite: | Dimerization of elongator protein 1 is essential for Elongator complex assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CQS
 
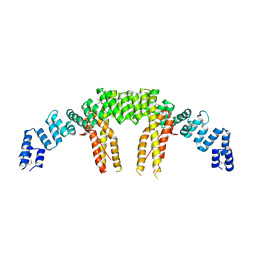 | | Dimerization of Elp1 is essential for Elongator complex assembly | | Descriptor: | Elongator complex protein 1 | | Authors: | Lin, Z, Xu, H, Li, F, Diao, W, Long, J, Shen, Y. | | Deposit date: | 2015-07-22 | | Release date: | 2015-08-19 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dimerization of elongator protein 1 is essential for Elongator complex assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6KIN
 
 | | Crystal structure of the tri-functional malyl-CoA lyase from Roseiflexus castenholzii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase | | Authors: | Tang, W.R, Zhang, C.Y, Wang, C, Xu, X.L. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.527 Å) | | Cite: | The C-terminal domain conformational switch revealed by the crystal structure of malyl-CoA lyase from Roseiflexus castenholzii.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
6ITQ
 
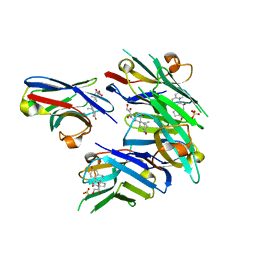 | | Crystal structure of cortisol complexed with its nanobody at pH 10.5 | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, SULFATE ION, anti-cortisol camelid antibody | | Authors: | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | Deposit date: | 2018-11-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
3RDZ
 
 | |
3RDY
 
 | |
8W9B
 
 | | CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an allosteric site | | Descriptor: | 1-[(1S)-1-(5-fluoranyl-3-methyl-1-benzofuran-2-yl)-2-methyl-propyl]-3-(1-oxidanylidene-2,3-dihydroisoindol-5-yl)urea, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Huang, X, Ren, X, Zhong, W. | | Deposit date: | 2023-09-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures reveal two allosteric inhibition modes of PI3K alpha H1047R involving a re-shaping of the activation loop.
Structure, 2024
|
|
8W9A
 
 | | CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-7909 binding at an allosteric site | | Descriptor: | 6-chloranyl-3-[[(1R)-1-[2-(1,3-dihydropyrrolo[3,4-c]pyridin-2-yl)-3,6-dimethyl-4-oxidanylidene-quinazolin-8-yl]ethyl]amino]pyridine-2-carboxylic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Huang, X, Ren, X, Zhong, W. | | Deposit date: | 2023-09-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures reveal two allosteric inhibition modes of PI3K alpha H1047R involving a re-shaping of the activation loop.
Structure, 2024
|
|
6ITP
 
 | | Crystal structure of cortisol complexed with its nanobody at pH 3.5 | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, anti-cortisol camelid antibody | | Authors: | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | Deposit date: | 2018-11-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
