7F41
 
 | |
7XAX
 
 | | Crystal structure of SARS coronavirus main protease in complex with Baicalei | | Descriptor: | 3C-like proteinase nsp5, 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one | | Authors: | Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2022-03-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of SARS-CoV 3C-like protease with baicalein.
Biochem.Biophys.Res.Commun., 611, 2022
|
|
7JI3
 
 | |
7VVP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7C2Y
 
 | | The crystal structure of COVID-2019 main protease in the apo state | | Descriptor: | 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zhou, H, Hu, X.H, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-05-10 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | COVID-2019 main protease in the apo state
To Be Published
|
|
7WQJ
 
 | | Crystal structure of MERS main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7XB3
 
 | |
7WQI
 
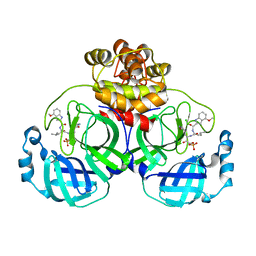 | | Crystal structure of SARS coronavirus main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of SARS coronavirus main protease in complex with PF07304814
To Be Published
|
|
7DR9
 
 | |
7YGQ
 
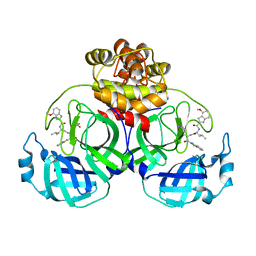 | | Crystal structure of SARS main protease in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-07-12 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7WQH
 
 | | Crystal structure of HCoV-NL63 main protease with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhong, F.L, Zhou, X.L, Lin, C, Zeng, P, Li, J, Zhang, J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7XRS
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Replicase polyprotein 1a | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-05-11 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7XRY
 
 | | Crystal structure of MERS main protease in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2022-05-12 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7E8L
 
 | | The structure of Spodoptera litura chemosensory protein | | Descriptor: | Putative chemosensory protein CSP8 | | Authors: | Xie, W, Jia, Q, Zeng, H, Xiao, N, Tang, J, Gao, S, Zhang, J. | | Deposit date: | 2021-03-02 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Spodoptera litura Chemosensory Protein CSP8.
Insects, 12, 2021
|
|
8J25
 
 | | Crystal structure of PML B-box2 mutant | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
8J2P
 
 | | Crystal structure of PML B-box2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
7F6M
 
 | | Crystal structure of APC complexed with a peptide inhibitor MAI-516 | | Descriptor: | Adenomatous polyposis coli protein, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Efficient Adenomatous Polyposis Coli - Rho Guanine Nucleotide Exchange Factor 4 Inhibitors by Employing High Binding Affinity Tracer
To Be Published
|
|
7F7O
 
 | | APC-Asef FP assay tracer | | Descriptor: | Adenomatous polyposis coli protein, N-[3',6'-bis(oxidanyl)-3-oxidanylidene-spiro[2-benzofuran-1,9'-xanthene]-5-yl]methanethioamide, Tracer 7 | | Authors: | Yang, X, Zhang, J. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The tracer of FP assay for screening APC-Asef inhbitors
To Be Published
|
|
7VBD
 
 | | Crystal structure of SARS-Cov-2 nucleocapsid N-terminal domain (NTD) protein,pH8.0 | | Descriptor: | Nucleoprotein | | Authors: | Zeng, P, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-08-31 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of SARS-Cov-2 nucleocapsid N-terminal domain (NTD) protein,pH8.0
To Be Published
|
|
7VBF
 
 | |
7VBE
 
 | |
7Y72
 
 | | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface) | | Descriptor: | Fab E7 heavy chain, Fab E7 light chain, Spike glycoprotein | | Authors: | Chia, W.N, Tan, C.W, Tan, A.W.K, Young, B, Starr, T.N, Lopez, E, Fibriansah, G, Barr, J, Cheng, S, Yeoh, A.Y.Y, Yap, W.C, Lim, B.L, Ng, T.S, Sia, W.R, Zhu, F, Chen, S, Zhang, J, Greaney, A.J, Chen, M, Au, G.G, Paradkar, P, Peiris, M, Chung, A.W, Bloom, J.D, Lye, D, Lok, S.M, Wang, L.F. | | Deposit date: | 2022-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Potent pan huACE2-dependent sarbecovirus neutralizing monoclonal antibodies isolated from a BNT162b2-vaccinated SARS survivor.
Sci Adv, 9, 2023
|
|
7Y71
 
 | | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab E7 heavy chain, ... | | Authors: | Chia, W.N, Tan, C.W, Tan, A.W.K, Young, B, Starr, T.N, Lopez, E, Fibriansah, G, Barr, J, Cheng, S, Yeoh, A.Y.Y, Yap, W.C, Lim, B.L, Ng, T.S, Sia, W.R, Zhu, F, Chen, S, Zhang, J, Greaney, A.J, Chen, M, Au, G.G, Paradkar, P, Peiris, M, Chung, A.W, Bloom, J.D, Lye, D, Lok, S.M, Wang, L.F. | | Deposit date: | 2022-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Potent pan huACE2-dependent sarbecovirus neutralizing monoclonal antibodies isolated from a BNT162b2-vaccinated SARS survivor.
Sci Adv, 9, 2023
|
|
7D6L
 
 | |
7DKA
 
 | | Crystal structure of DsbA-like protein DR2335 from Deinococcus radiodurans R1, C24S mutant protein | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, M.-K, Zhang, J, Zhao, L. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of DsbA-like protein DR2335 from Deinococcus radiodurans R1, native protein
To Be Published
|
|
