2CCC
 
 | | Complexes of Dodecin with Flavin and Flavin-like Ligands | | Descriptor: | CHLORIDE ION, LUMIFLAVIN, MAGNESIUM ION, ... | | Authors: | Grininger, M, Zeth, K, Oesterhelt, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dodecins: A Family of Lumichrome Binding Proteins.
J.Mol.Biol., 357, 2006
|
|
2CO5
 
 | | F93 FROM STIV, a winged-helix DNA-binding protein | | Descriptor: | VIRAL PROTEIN F93 | | Authors: | Larson, E.T, Reiter, D, Lawrence, C.M. | | Deposit date: | 2006-05-25 | | Release date: | 2006-05-31 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Winged-Helix Protein from Sulfolobus Turreted Icosahedral Virus Points Toward Stabilizing Disulfide Bonds in the Intracellular Proteins of a Hyperthermophilic Virus.
Virology, 368, 2007
|
|
8UYH
 
 | | Structure of AMP-PNP-bound Pediculus humanus (Ph) PINK1 dimer | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Pink1, ... | | Authors: | Gan, Z.Y, Kirk, N.S, Leis, A, Komander, D. | | Deposit date: | 2023-11-13 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Interaction of PINK1 with nucleotides and kinetin.
Sci Adv, 10, 2024
|
|
2CI7
 
 | | Crystal structure of Dimethylarginine Dimethylaminohydrolase I in complex with Zinc, high pH | | Descriptor: | GLYCINE, NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1, ... | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inhibitors.
Structure, 14, 2006
|
|
2CGL
 
 | |
2CIR
 
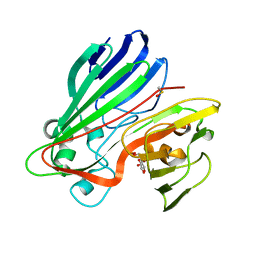 | | Structure-based functional annotation: Yeast ymr099c codes for a D- hexose-6-phosphate mutarotase. Complex with glucose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, HEXOSE-6-PHOSPHATE MUTAROTASE | | Authors: | Graille, M, Baltaze, J.-P, Leulliot, N, Liger, D, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2006-03-24 | | Release date: | 2006-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based functional annotation: yeast ymr099c codes for a D-hexose-6-phosphate mutarotase.
J. Biol. Chem., 281, 2006
|
|
2CIC
 
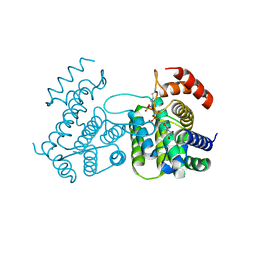 | | THE CRYSTAL STRUCTURE OF A COMPLEX OF CAMPYLOBACTER JEJUNI DUTPASE WITH SUBSTRATE ANALOGUE DUPNHPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDE HYDROLASE, MAGNESIUM ION | | Authors: | Moroz, O.V, Harkiolaki, M, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2006-03-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the Leishmania Major Deoxyuridine Triphosphate Nucleotidohydrolase in Complex with Nucleotide Analogues, Dump, and Deoxyuridine.
J.Biol.Chem., 286, 2011
|
|
2CI6
 
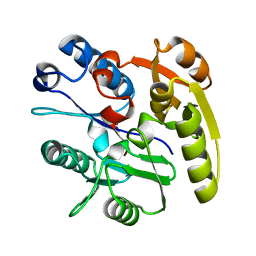 | | Crystal Structure of Dimethylarginine dimethylaminohydrolase I bound with Zinc low pH | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1, ZINC ION | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inbitors
Structure, 14, 2006
|
|
3C1J
 
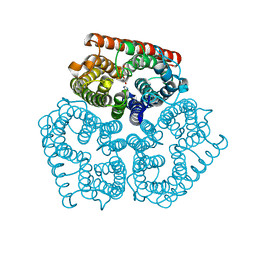 | | Substrate binding, deprotonation and selectivity at the periplasmic entrance of the E. coli ammonia channel AmtB | | Descriptor: | ACETATE ION, Ammonia channel, GLYCEROL, ... | | Authors: | Lupo, D, Winkler, F.K. | | Deposit date: | 2008-01-23 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate binding, deprotonation, and selectivity at the periplasmic entrance of the Escherichia coli ammonia channel AmtB.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2CHF
 
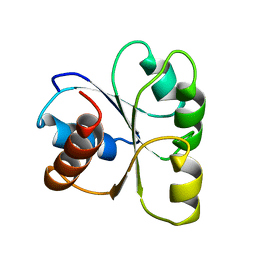 | | STRUCTURE OF THE MG2+-BOUND FORM OF CHEY AND THE MECHANISM OF PHOSPHORYL TRANSFER IN BACTERIAL CHEMOTAXIS | | Descriptor: | CHEY | | Authors: | Stock, A, Martinez-Hackert, E, Rasmussen, B, West, A, Stock, J, Ringe, D, Petsko, G. | | Deposit date: | 1994-01-17 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis.
Biochemistry, 32, 1993
|
|
2CLE
 
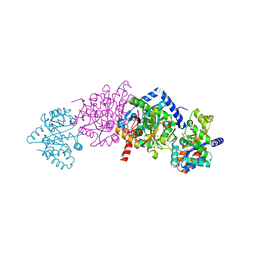 | | Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2- amino-1-ethylphosphate (F6) - lowF6 complex | | Descriptor: | 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and Characterization of Allosteric Probes of Substrate Channeling in the Tryptophan Synthase Bienzyme Complex.
Biochemistry, 46, 2007
|
|
2CHM
 
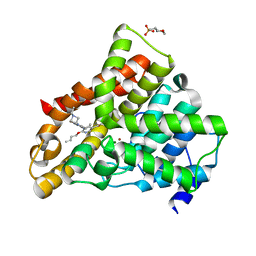 | | Crystal structure of N2 substituted pyrazolo pyrimidinones - a flipped binding mode in PDE5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[2-(BUT-3-EN-1-YLOXY)-5-(1-HYDROXYVINYL)PYRIDIN-3-YL]-3-ETHYL-2-(1-ETHYLAZETIDIN-3-YL)-1,2,6,7A-TETRAHYDRO-7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, CGMP-SPECIFIC 3', ... | | Authors: | Allerton, C.M.N, Barber, C.G, Beaumont, K.C, Brown, D.G, Cole, S.M, Ellis, D, Lane, C.A.L, Maw, G.N, Mount, N.M, Rawson, D.J, Robinson, C.M, Street, S.D.A, Summerhill, N.W. | | Deposit date: | 2006-03-15 | | Release date: | 2006-06-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Novel Series of Potent and Selective Pde5 Inhibitors with Potential for High and Dose-Independent Oral Bioavailability
J.Med.Chem., 49, 2006
|
|
2C7O
 
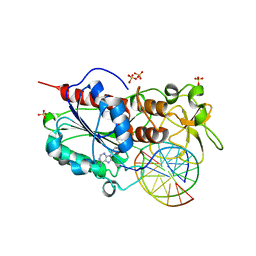 | |
8VSA
 
 | | Endogenous trans-translation complex with tmRNA*SmpB in the P site and alanyl-tRNA in the A site of E. coli 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Teran, D, Zhang, Y, Korostelev, A.A. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Endogenous trans-translation structure visualizes the decoding of the first tmRNA alanine codon.
Front Microbiol, 15, 2024
|
|
7U0S
 
 | | Crystal Structure of FK506-binding protein 1A from Aspergillus fumigatus Bound to Ascomycin | | Descriptor: | (3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,22R,26aS)-8-ethyl-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-5,6,8,11,12,13,14,15,16,17,18,19,24,25,26,26a-hexadecahydro-3H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosine-1,7,20,21(4H,23H)-tetrone, ACETATE ION, FK506-binding protein 1A, ... | | Authors: | DeBouver, N.D, Fox III, D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
3C5I
 
 | | Crystal structure of Plasmodium knowlesi choline kinase, PKH_134520 | | Descriptor: | CALCIUM ION, CHOLINE ION, Choline kinase, ... | | Authors: | Wernimont, A.K, Hills, T, Lew, J, Wasney, G, Senesterra, G, Kozieradzki, I, Cossar, D, Vedadi, M, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Plasmodium knowlesi choline kinase, PKH_134520.
To be Published
|
|
3C75
 
 | | Paracoccus versutus methylamine dehydrogenase in complex with amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION, Methylamine dehydrogenase heavy chain, ... | | Authors: | Cavalieri, C, Biermann, N, Vlasie, M.D, Einsle, O, Merli, A, Ferrari, D, Rossi, G.L, Ubbink, M. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural comparison of crystal and solution states of the 138 kDa complex of methylamine dehydrogenase and amicyanin from Paracoccus versutus.
Biochemistry, 47, 2008
|
|
8VS9
 
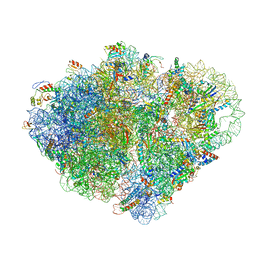 | | Endogenous trans-translation complex with tmRNA*SmpB in the P site and alanyl-tRNA in the A site and deacyl-tRNA in the E site of E. coli 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Teran, D, Zhang, Y, Korostelev, A.A. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Endogenous trans-translation structure visualizes the decoding of the first tmRNA alanine codon.
Front Microbiol, 15, 2024
|
|
7U0T
 
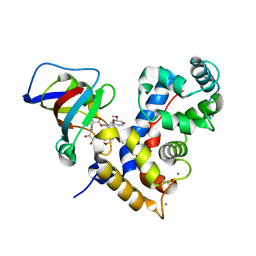 | | Crystal Structure of a human Calcineurin A - Calcineurin B fusion bound to FKBP12 and FK-520 | | Descriptor: | (3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,22R,26aS)-8-ethyl-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-5,6,8,11,12,13,14,15,16,17,18,19,24,25,26,26a-hexadecahydro-3H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosine-1,7,20,21(4H,23H)-tetrone, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Fox III, D, Mayclin, S.J, DeBouver, N.D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
7U0U
 
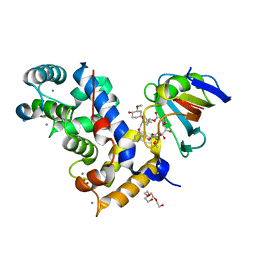 | | Crystal Structure of a Aspergillus fumigatus Calcineurin A - Calcineurin B fusion bound to FKBP12 and FK-506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Fox III, D, Abendroth, J, DeBouver, N.D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
2CLS
 
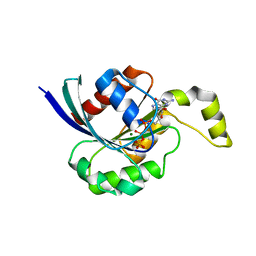 | | The crystal structure of the human RND1 GTPase in the active GTP bound state | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RHO-RELATED GTP-BINDING PROTEIN RHO6 | | Authors: | Pike, A.C.W, Yang, X, Colebrook, S, Gileadi, O, Sobott, F, Bray, J, Wen Hwa, L, Marsden, B, Zhao, Y, Schoch, G, Elkins, J, Debreczeni, J.E, Turnbull, A.P, von Delft, F, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Crystal Structure of the Human Rnd1 Gtpase in the Active GTP Bound State
To be Published
|
|
8WN8
 
 | | CryoEM structure of ZIKV rsNS1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Non-structural protein 1 | | Authors: | Chew, B.L.A, Luo, D. | | Deposit date: | 2023-10-05 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of Zika virus NS1 multimerization and human antibody recognition
Npj Viruses, 2, 2024
|
|
3AVU
 
 | | Structure of viral RNA polymerase complex 2 | | Descriptor: | CALCIUM ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
8WNP
 
 | |
2CLK
 
 | | Tryptophan Synthase in complex with D-glyceraldehyde 3-phosphate (G3P) | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and Characterization of Allosteric Probes of Substrate Channeling in the Tryptophan Synthase Bienzyme Complex.
Biochemistry, 46, 2007
|
|
