8ZFL
 
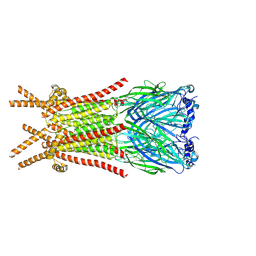 | | Caenorhabditis elegans ACR-23 in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betaine receptor acr-23,Soluble cytochrome b562 | | Authors: | Chen, Q.F, Liu, F.L, Li, T.Y, Gong, H.H, Guo, F, Liu, S. | | Deposit date: | 2024-05-08 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural insights into the molecular effects of the anthelmintics monepantel and betaine on the Caenorhabditis elegans acetylcholine receptor ACR-23.
Embo J., 43, 2024
|
|
8ZFM
 
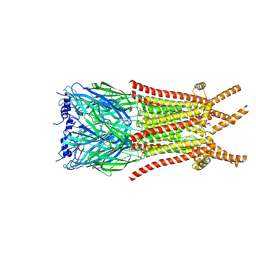 | | Caenorhabditis elegans ACR-23 in betaine bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betaine receptor acr-23,Soluble cytochrome b562, ... | | Authors: | Chen, Q.F, Liu, F.L, Li, T.Y, Gong, H.H, Guo, F, Liu, S. | | Deposit date: | 2024-05-08 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural insights into the molecular effects of the anthelmintics monepantel and betaine on the Caenorhabditis elegans acetylcholine receptor ACR-23.
Embo J., 43, 2024
|
|
8ZEM
 
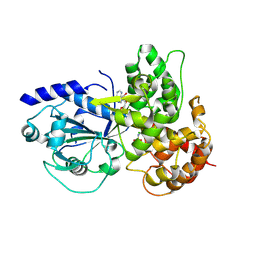 | | Crystal Structure of NLRP3 NACHT domain in complex with NP3-1 | | Descriptor: | 1-[5-[2,3-bis(chloranyl)phenyl]-2,3-dihydro-1~{H}-inden-4-yl]-3-[4-(2-oxidanylpropan-2-yl)thiophen-2-yl]sulfonyl-urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Shi, C, Liu, Z.M. | | Deposit date: | 2024-05-06 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Deep-Learning-Driven Discovery of SN3-1, a Potent NLRP3 Inhibitor with Therapeutic Potential for Inflammatory Diseases.
J.Med.Chem., 67, 2024
|
|
8X6B
 
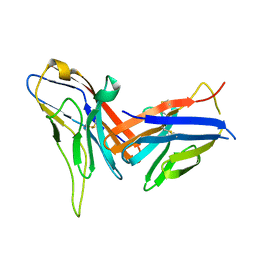 | | Crystal structure of immune receptor PVRIG in complex with ligand Nectin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nectin-2, Transmembrane protein PVRIG | | Authors: | Hu, S.T, Han, P, Wang, H, Qi, J.X. | | Deposit date: | 2023-11-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the immune recognition and selectivity of the immune receptor PVRIG for ligand Nectin-2.
Structure, 32, 2024
|
|
8ZFK
 
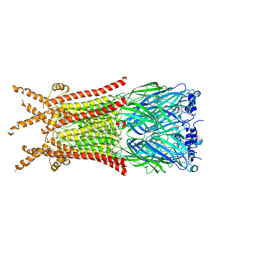 | | Caenorhabditis elegans ACR-23 in betaine and monepantel bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betaine receptor acr-23,Soluble cytochrome b562, ... | | Authors: | Chen, Q.F, Liu, F.L, Li, T.Y, Gong, H.H, Guo, F, Liu, S. | | Deposit date: | 2024-05-08 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural insights into the molecular effects of the anthelmintics monepantel and betaine on the Caenorhabditis elegans acetylcholine receptor ACR-23.
Embo J., 43, 2024
|
|
8K2W
 
 | | Structure of CXCR3 complexed with antagonist AMG487 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, N-[(1R)-1-[3-(4-ethoxyphenyl)-4-oxidanylidene-pyrido[2,3-d]pyrimidin-2-yl]ethyl]-N-(pyridin-3-ylmethyl)-2-[4-(trifluoromethyloxy)phenyl]ethanamide, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure basis for the modulation of CXC chemokine receptor 3 by antagonist AMG487.
Cell Discov, 9, 2023
|
|
8JP0
 
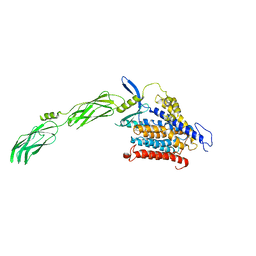 | | structure of human sodium-calciumexchanger NCX1 | | Descriptor: | 2-{4-[(2,5-difluorophenyl)methoxy]phenoxy}-5-ethoxyaniline, Sodium/calcium exchanger 1 | | Authors: | Dong, Y, Zhao, Y. | | Deposit date: | 2023-06-09 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insight into the allosteric inhibition of human sodium-calcium exchanger NCX1 by XIP and SEA0400.
Embo J., 43, 2024
|
|
8K2X
 
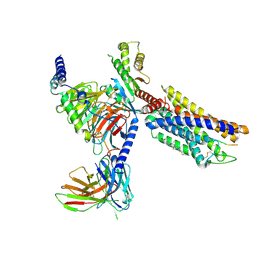 | | CXCR3-DNGi complex activated by CXCL10 | | Descriptor: | C-X-C motif chemokine 10, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure basis for the modulation of CXC chemokine receptor 3 by antagonist AMG487.
Cell Discov, 9, 2023
|
|
7JR7
 
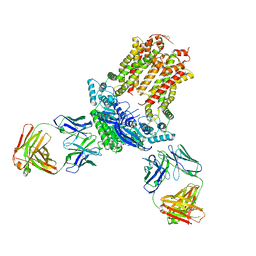 | | Cryo-EM structure of ABCG5/G8 in complex with Fab 2E10 and 11F4 | | Descriptor: | ATP-binding cassette sub-family G member 5, ATP-binding cassette sub-family G member 8, Fab 11F4 heavy chain, ... | | Authors: | Huang, C.S, Yu, X, Min, X, Wang, Z, Zhang, H. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of ABCG5/G8 in complex with modulating antibodies
Commun Biol, 4, 2021
|
|
7DOQ
 
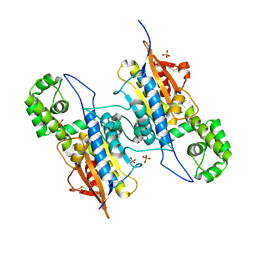 | | Lp major histidine acid phosphatase mutant D281A/5'-AMP | | Descriptor: | Acid phosphatase, PHOSPHATE ION | | Authors: | Guo, Y, Teng, Y. | | Deposit date: | 2020-12-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into a new substrate binding mode of a histidine acid phosphatase from Legionella pneumophila.
Biochem.Biophys.Res.Commun., 540, 2021
|
|
8I6V
 
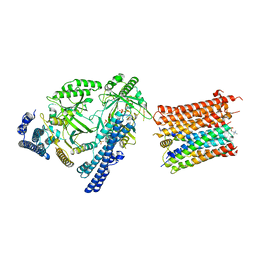 | | Cryo-EM structure of the polyphosphate polymerase VTC complex(Vtc4/Vtc3/Vtc1) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Mayer, A, Wu, S, Ye, S. | | Deposit date: | 2023-01-29 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structure of the polyphosphate polymerase VTC reveals coupling of polymer synthesis to membrane transit.
Embo J., 42, 2023
|
|
8I8Y
 
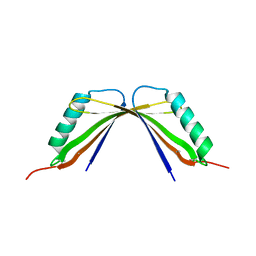 | | A mutant of the C-terminal complex of proteins 4.1G and NuMA | | Descriptor: | Engineered protein | | Authors: | Hu, X. | | Deposit date: | 2023-02-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Combined prediction and design reveals the target recognition mechanism of an intrinsically disordered protein interaction domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7D2F
 
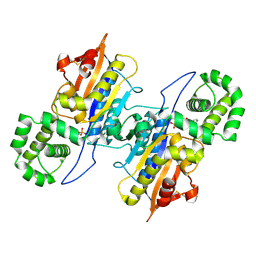 | | Lp major histidine acid phosphatase mutant D281A/5'-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Major acid phosphatase | | Authors: | Guo, Y, Teng, Y. | | Deposit date: | 2020-09-16 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into a new substrate binding mode of a histidine acid phosphatase from Legionella pneumophila.
Biochem.Biophys.Res.Commun., 540, 2021
|
|
7CAB
 
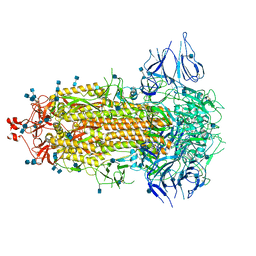 | | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CYP
 
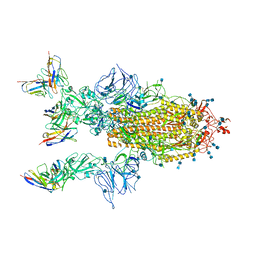 | | Complex of SARS-CoV-2 spike trimer with its neutralizing antibody HB27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of HB27, ... | | Authors: | Wang, X, Zhu, L. | | Deposit date: | 2020-09-04 | | Release date: | 2021-06-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Double lock of a potent human therapeutic monoclonal antibody against SARS-CoV-2.
Natl Sci Rev, 8, 2021
|
|
7CAC
 
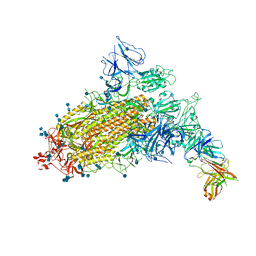 | | SARS-CoV-2 S trimer with one RBD in the open state and complexed with one H014 Fab. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7C96
 
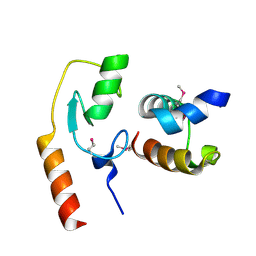 | | Avr1d:GmPUB13 U-box | | Descriptor: | RING-type E3 ubiquitin transferase, RxLR effector protein Avh6 | | Authors: | Xing, W, Hu, Q, Zhou, J, Yao, D. | | Deposit date: | 2020-06-05 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CAI
 
 | | SARS-CoV-2 S trimer with two RBDs in the open state and complexed with two H014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAH
 
 | | The interface of H014 Fab binds to SARS-CoV-2 S | | Descriptor: | Heavy chain of H014 Fab, Light chain of H014 Fab, Spike protein S1 | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Rao, Z, wang, Y, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAK
 
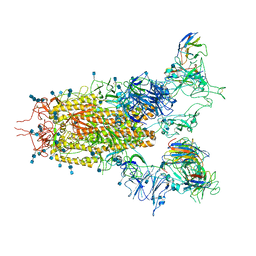 | | SARS-CoV-2 S trimer with three RBD in the open state and complexed with three H014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7YTX
 
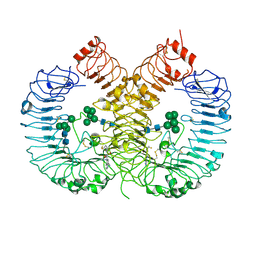 | | Crystal structure of TLR8 in complex with its antagonist | | Descriptor: | (2R,6R)-4-(8-cyanoquinolin-5-yl)-N-[(3S,4R)-4-fluoranylpyrrolidin-3-yl]-6-methyl-morpholine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shimizu, T, Sakaniwa, K. | | Deposit date: | 2022-08-16 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A novel Toll-like receptor 7/8-specific antagonist E6742 ameliorates clinically relevant disease parameters in murine models of lupus.
Eur.J.Pharmacol., 957, 2023
|
|
7YTP
 
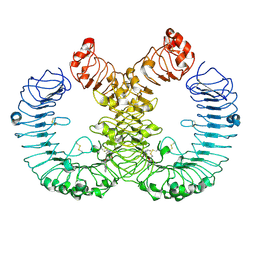 | | TLR7 in complex with an inhibitor | | Descriptor: | (2R,6R)-4-(8-cyanoquinolin-5-yl)-N-[(3S,4R)-4-fluoranylpyrrolidin-3-yl]-6-methyl-morpholine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2022-08-15 | | Release date: | 2023-11-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | A novel Toll-like receptor 7/8-specific antagonist E6742 ameliorates clinically relevant disease parameters in murine models of lupus.
Eur.J.Pharmacol., 957, 2023
|
|
7Y63
 
 | | ApoSIDT2-pH7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2, ... | | Authors: | Gong, D.S. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insight into the human SID1 transmembrane family member 2 reveals its lipid hydrolytic activity.
Nat Commun, 14, 2023
|
|
7Y68
 
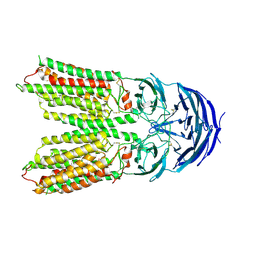 | | SIDT2-pH5.5 plus miRNA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2, ... | | Authors: | Gong, D.S. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insight into the human SID1 transmembrane family member 2 reveals its lipid hydrolytic activity.
Nat Commun, 14, 2023
|
|
7Y69
 
 | | ApoSIDT2-pH5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2, ... | | Authors: | Gong, D.S. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural insight into the human SID1 transmembrane family member 2 reveals its lipid hydrolytic activity.
Nat Commun, 14, 2023
|
|
