7QMX
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 7) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QN0
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 10) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QN2
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 12) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7YXW
 
 | | Structure of the p22phox A200G mutant in complex with p47phox peptide | | Descriptor: | Cytochrome b-245 light chain, Neutrophil cytosol factor 1 | | Authors: | Cukier, C.D, Vuillard, L.M, Komjati, B, Szlavik, Z. | | Deposit date: | 2022-02-16 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting NOX2 via p47/phox-p22/phox Inhibition with Novel Triproline Mimetics
Acs Med.Chem.Lett., 13, 2022
|
|
5EDS
 
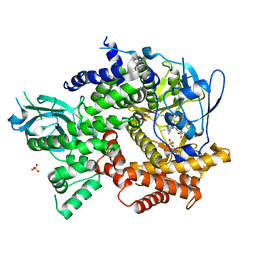 | | Crystal structure of human PI3K-gamma in complex with benzimidazole inhibitor 5 | | Descriptor: | 4-azanyl-6-[[(1~{S})-1-[6-fluoranyl-1-(3-methylsulfonylphenyl)benzimidazol-2-yl]ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery, Optimization, and in Vivo Evaluation of Benzimidazole Derivatives AM-8508 and AM-9635 as Potent and Selective PI3K delta Inhibitors.
J.Med.Chem., 59, 2016
|
|
7YX9
 
 | |
7YXB
 
 | | MHC-II dynamics are maintained in HLA-DR allotypes to ensure catalyzed peptide exchange | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, CLIP peptide, ... | | Authors: | Roske, Y, Abualrous, E.T, Freund, C. | | Deposit date: | 2022-02-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | MHC-II dynamics are maintained in HLA-DR allotypes to ensure catalyzed peptide exchange.
Nat.Chem.Biol., 19, 2023
|
|
7Z0Q
 
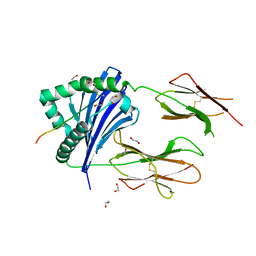 | | MHC-II dynamics are maintained in HLA-DR allotypes to ensure catalyzed peptide exchange | | Descriptor: | 1,2-ETHANEDIOL, CLIP peptide, HLA class II histocompatibility antigen, ... | | Authors: | Roske, Y, Abualrous, E.T, Freund, C. | | Deposit date: | 2022-02-23 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MHC-II dynamics are maintained in HLA-DR allotypes to ensure catalyzed peptide exchange.
Nat.Chem.Biol., 19, 2023
|
|
8A46
 
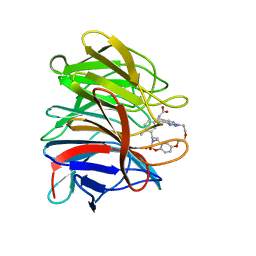 | | Crystal structure of the human Kelch domain of Keap1 in complex with compound S217879 | | Descriptor: | 2-[(1S,2R,8S)-2,4,32-trimethyl-28,28-bis(oxidanylidene)-19,22,27-trioxa-28$l^{6}-thia-1,14,15,16-tetrazahexacyclo[21.5.3.1^{3,7}.1^{9,13}.0^{12,16}.0^{26,30}]tritriaconta-3(33),4,6,9(32),10,12,14,23,25,30-decaen-8-yl]ethanoic acid, Kelch-like ECH-associated protein 1 | | Authors: | Weber, C, Vuillard, L, Delerive, P, Miallau, L. | | Deposit date: | 2022-06-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.323 Å) | | Cite: | Selective disruption of NRF2-KEAP1 interaction leads to NASH resolution and reduction of liver fibrosis in mice.
JHEP Rep, 5, 2023
|
|
8RWL
 
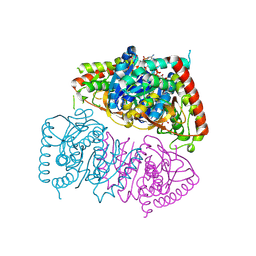 | | Crystal structure of Methanopyrus kandleri malate dehydrogenase mutant 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Malate dehydrogenase, ... | | Authors: | Coquille, S, Roche, J, Engilberge, S, Girard, E, Madern, D. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Navigating the conformational landscape of an enzyme. Stabilization of a low populated conformer by evolutionary mutations triggers Allostery into a non-allosteric enzyme.
To Be Published
|
|
8RS5
 
 | |
4IRJ
 
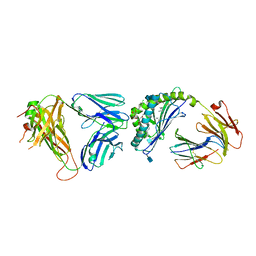 | | Structure of the mouse CD1d-4ClPhC-alpha-GalCer-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Nemcovic, M, Zajonc, D.M. | | Deposit date: | 2013-01-14 | | Release date: | 2013-08-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Enhanced TCR footprint by a novel glycolipid increases NKT-dependent tumor protection.
J.Immunol., 191, 2013
|
|
4IRS
 
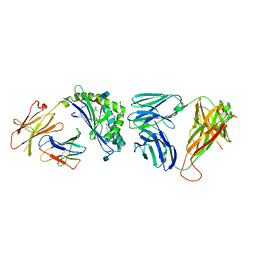 | | Structure of the mouse CD1d-PyrC-alpha-GalCer-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nemcovic, M, Zajonc, D.M. | | Deposit date: | 2013-01-15 | | Release date: | 2013-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enhanced TCR footprint by a novel glycolipid increases NKT-dependent tumor protection.
J.Immunol., 191, 2013
|
|
7RNI
 
 | |
7RN5
 
 | |
6OOR
 
 | | Structure of 1B1 bound to mouse CD1d | | Descriptor: | Antibody 1B1 Heavy chain, Antibody 1B1 Light chain, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Ying, G, Zajonc, D.M. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of NKT cell inhibition using the T-cell receptor-blocking anti-CD1d antibody 1B1.
J.Biol.Chem., 294, 2019
|
|
1HAV
 
 | | HEPATITIS A VIRUS 3C PROTEINASE | | Descriptor: | CHLORIDE ION, HEPATITIS A VIRUS 3C PROTEINASE | | Authors: | Bergmann, E.M, James, M.N.G. | | Deposit date: | 1996-10-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition.
J.Virol., 71, 1997
|
|
7R1E
 
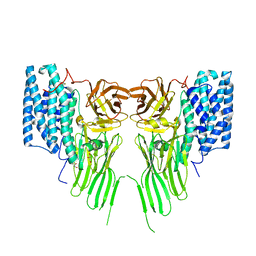 | | Mosquitocidal Cry11Ba determined at pH 10.4 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | GLYCEROL, Pesticidal crystal protein Cry11Ba | | Authors: | Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
6OJP
 
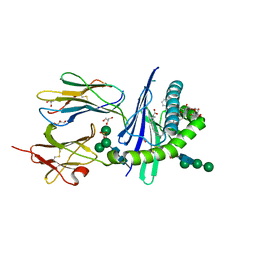 | | Structure of glycolipid alpha-GSA[8,6P] in complex with mouse CD1d | | Descriptor: | (5R,6S,7S)-5,6-dihydroxy-7-(octanoylamino)-N-(6-phenylhexyl)-8-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]oxy}octanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A. | | Deposit date: | 2019-04-11 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
7QYD
 
 | | mosquitocidal Cry11Ba determined at pH 6.5 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Ba | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX4
 
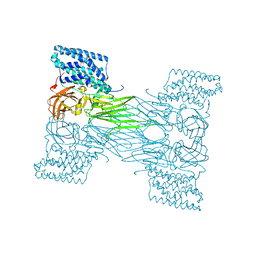 | | mosquitocidal Cry11Aa determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX6
 
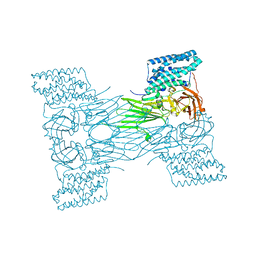 | | mosquitocidal Cry11Aa-E583Q determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX5
 
 | | mosquitocidal Cry11Aa-Y449F determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX7
 
 | | mosquitocidal Cry11Aa-F17Y determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7PI6
 
 | | Trypanosoma brucei ISG65 bound to human complement C3d | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 65 kDa invariant surface glycoprotein, Complement C3dg fragment, ... | | Authors: | Cook, A.D, Higgins, M.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Invariant surface glycoprotein 65 of Trypanosoma brucei is a complement C3 receptor.
Nat Commun, 13, 2022
|
|
