3RER
 
 | | Crystal structure of E. coli Hfq in complex with AU6A RNA and ADP | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP*UP*UP*A)-3', ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, W.W, Wu, J.H, Shi, Y.Y. | | Deposit date: | 2011-04-05 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cooperation of Escherichia coli Hfq hexamers in DsrA binding.
Genes Dev., 25, 2011
|
|
3RES
 
 | |
7FAS
 
 | | VAR2CSA 3D7 ectodomain core region | | Descriptor: | Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Zhaoning, W. | | Deposit date: | 2021-07-07 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
3SX4
 
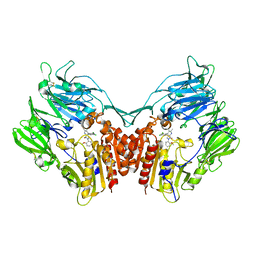 | | Crystal structure of human dpp-iv in complex with sa-(+)-3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyphenyl)- 2-methyl-5h-pyrrolo[3,4-b]pyridin-7(6h)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyphenyl)-2-methyl-5,6-dihydro-7H-pyrrolo[3,4-b]pyridin-7-one, ... | | Authors: | Klei, H.E. | | Deposit date: | 2011-07-14 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 7-Oxopyrrolopyridine-derived DPP4 inhibitors-mitigation of CYP and hERG liabilities via introduction of polar functionalities in the active site.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SWW
 
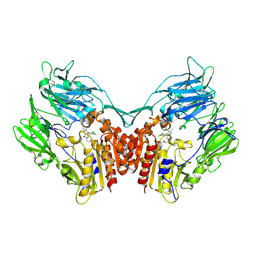 | | Crystal structure of human dpp-iv in complex with sa-(+)-3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyphenyl)- 2-methyl-5h-pyrrolo[3,4-b]pyridin-7(6h)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyethyl)-2-methyl-5,6-dihydro-7H-pyrrolo[3,4-b]pyridin-7-one, ... | | Authors: | Klei, H.E. | | Deposit date: | 2011-07-14 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 7-Oxopyrrolopyridine-derived DPP4 inhibitors-mitigation of CYP and hERG liabilities via introduction of polar functionalities in the active site.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7XZ4
 
 | |
7FAP
 
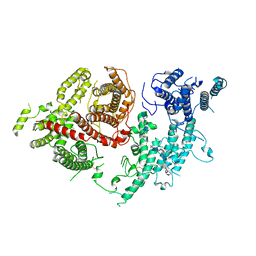 | | Structure of VAR2CSA-CSA 3D7 | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-3)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Wang, Z. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
5HAD
 
 | |
6HHO
 
 | | Crystal structure of RIP1 kinase in complex with GSK547 | | Descriptor: | 6-[4-[(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]carbonylpiperidin-1-yl]pyrimidine-4-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer.
Cancer Cell, 34, 2018
|
|
8XZG
 
 | | Cryo-EM structure of the [Pyr1]-apelin-13-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Apelin-13, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZF
 
 | | Cryo-EM structure of the WN561-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZJ
 
 | | Cryo-EM structure of the WN353-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZI
 
 | | Cryo-EM structure of the CMF-019-bound human APLNR-Gi complex | | Descriptor: | (3~{S})-5-methyl-3-[[1-pentan-3-yl-2-(thiophen-2-ylmethyl)benzimidazol-5-yl]carbonylamino]hexanoic acid, Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZH
 
 | | Cryo-EM structure of the MM07-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
3CTB
 
 | | Tethered PXR-LBD/SRC-1p apoprotein | | Descriptor: | Pregnane X receptor, Linker, Steroid receptor coactivator 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2008-04-11 | | Release date: | 2008-12-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Construction and characterization of a fully active PXR/SRC-1 tethered protein with increased stability
Protein Eng.Des.Sel., 21, 2008
|
|
7DIE
 
 | | Crystal structure of M. penetrans Ferritin | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, w.m, Zhang, y, Wang, h.f. | | Deposit date: | 2020-11-19 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ferritin with Atypical Ferroxidase Centers Takes B-Channels as the Pathway for Fe 2+ Uptake from Mycoplasma .
Inorg.Chem., 60, 2021
|
|
7WRG
 
 | | Crystal structure of full-length kinesin-3 KLP-6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein, MAGNESIUM ION | | Authors: | Wang, W.J, Ren, J.Q, Song, W.Y, Feng, W. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | The architecture of kinesin-3 KLP-6 reveals a multilevel-lockdown mechanism for autoinhibition.
Nat Commun, 13, 2022
|
|
6LGN
 
 | | The atomic structure of varicella zoster virus C-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Li, S, Zheng, Q. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
8JMX
 
 | |
4EFG
 
 | |
4EDE
 
 | |
4EEJ
 
 | |
7DWO
 
 | |
7DWN
 
 | | Crystal structure of Vibrio fischeri DarR in complex with DNA reveals the transcriptional activation mechanism of LTTR family members | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Predicted DNA-binding transcriptional regulator | | Authors: | Wang, W.W, Wu, H, He, J.H, Yu, F. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure details of Vibrio fischeri DarR and mutant DarR-M202I from LTTR family reveals their activation mechanism.
Int.J.Biol.Macromol., 183, 2021
|
|
