1I87
 
 | | SOLUTION STRUCTURE OF THE WATER-SOLUBLE FRAGMENT OF RAT HEPATIC APOCYTOCHROME B5 | | Descriptor: | CYTOCHROME B5 | | Authors: | Falzone, C.J, Wang, Y, Vu, B.C, Scott, N.L, Bhattacharya, S, Lecomte, J.T. | | Deposit date: | 2001-03-12 | | Release date: | 2001-05-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic perturbations induced by heme binding in cytochrome b5.
Biochemistry, 40, 2001
|
|
3HHS
 
 | | Crystal Structure of Manduca sexta prophenoloxidase | | Descriptor: | COPPER (II) ION, Phenoloxidase subunit 1, Phenoloxidase subunit 2 | | Authors: | Li, Y, Wang, Y, Jiang, H, Deng, J. | | Deposit date: | 2009-05-17 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of Manduca sexta prophenoloxidase provides insights into the mechanism of type 3 copper enzymes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6P8D
 
 | |
6PEF
 
 | |
6PEC
 
 | |
6PC4
 
 | | Tubulin-RB3_SLD-TTL in complex with compound ABI-274 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structure-Activity Relationship Study of Novel 6-Aryl-2-benzoyl-pyridines as Tubulin Polymerization Inhibitors with Potent Antiproliferative Properties.
J.Med.Chem., 63, 2020
|
|
5KJS
 
 | | Crystal Structure of Arabidopsis thaliana HCT | | Descriptor: | Shikimate O-hydroxycinnamoyltransferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
6PDS
 
 | | Vaccine-elicited NHP FP-targeting antibody 0PV-a.04 in complex with HIV fusion peptide (residue 512-519) | | Descriptor: | HIV-1 fusion peptide residue 512-519, SULFATE ION, antibody 0PV-a.04 heavy chain, ... | | Authors: | Xu, K, Liu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Modular recognition of antigens provides a mechanism that improves vaccine-elicited antibody-class frequencies
To Be Published
|
|
4BGG
 
 | | Crystal structure of the ACVR1 kinase in complex with LDN-213844 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenyl}piperazine, ACTIVIN RECEPTOR TYPE-1, ... | | Authors: | Sanvitale, C, Canning, P, Cooper, C, Wang, Y, Mohedas, A.H, Choi, S, Yu, P.B, Cuny, G.D, Nowak, R, Coutandin, D, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-26 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure-Activity Relationship of 3,5-Diaryl-2-Aminopyridine Alk2 Inhibitors Reveals Unaltered Binding Affinity for Fibrodysplasia Ossificans Progressiva Causing Mutants.
J.Med.Chem., 57, 2014
|
|
6D88
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 13f | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2018-04-26 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structural Modification of the 3,4,5-Trimethoxyphenyl Moiety in the Tubulin Inhibitor VERU-111 Leads to Improved Antiproliferative Activities.
J. Med. Chem., 61, 2018
|
|
6PDR
 
 | |
6PDU
 
 | | Vaccine-elicited NHP FP-targeting antibody 13N024-a.01 in complex with HIV fusion peptide (residue 512-519) | | Descriptor: | HIV-1 fusion peptide residue 512-519, antibody 13N024-a.01, heavy chain, ... | | Authors: | Xu, K, Liu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Modular recognition of antigens provides a mechanism that improves vaccine-elicited antibody-class frequencies
To Be Published
|
|
5KJT
 
 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroyl-CoA | | Descriptor: | Shikimate O-hydroxycinnamoyltransferase, p-coumaroyl-CoA | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJW
 
 | | Crystal structure of Coleus blumei HCT in complex with 3-hydroxyacetophenone | | Descriptor: | 1-(3-hydroxyphenyl)ethanone, Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
4MGR
 
 | | The crystal structure of Bacillus subtilis GabR, an autorepressor and PLP- and GABA-dependent transcriptional activator of gabT | | Descriptor: | ACETATE ION, HTH-type transcriptional regulatory protein GabR, IMIDAZOLE, ... | | Authors: | Wu, R, Edayathumangalam, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6QGK
 
 | | Structure of human Bcl-2 in complex with THIQ-phenyl pyrazole compound | | Descriptor: | 1-[2-[[(3~{S})-3-(aminomethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]phenyl]-~{N},~{N}-dibutyl-5-methyl-pyrazole-3-carboxamide, ACETATE ION, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QGD
 
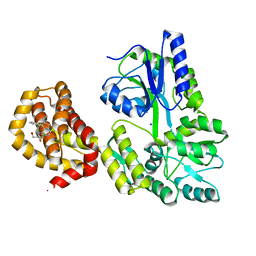 | | Structure of human Mcl-1 in complex with thienopyrimidine inhibitor | | Descriptor: | 2-[(6-ethyl-5-phenyl-thieno[2,3-d]pyrimidin-4-yl)amino]-3-oxidanyl-propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QGG
 
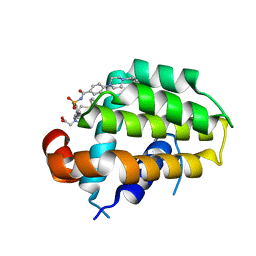 | | Structure of human Bcl-2 in complex with analogue of ABT-737 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1, [(3~{R})-3-[[4-[[4-[4-[[2-(4-chlorophenyl)phenyl]methyl]piperazin-1-yl]phenyl]carbonylsulfamoyl]-2-nitro-phenyl]amino]-4-phenylsulfanyl-butyl]-(2-hydroxy-2-oxoethyl)-dimethyl-azanium | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
2YJY
 
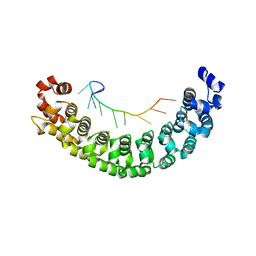 | | A specific and modular binding code for cytosine recognition in PUF domains | | Descriptor: | 5'-R(*AP*UP*UP*GP*CP*AP*UP*AP*UP*AP)-3', PUMILIO HOMOLOG 1 | | Authors: | Dong, S, Wang, Y, Cassidy-Amstutz, C, Lu, G, Qiu, C, Bigler, R, Jezyk, M, Li, C, Hall, T.M.T, Wang, Z. | | Deposit date: | 2011-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Specific and Modular Binding Code for Cytosine Recognition in Pumilio/Fbf (Puf) RNA-Binding Domains.
J.Biol.Chem., 286, 2011
|
|
6P7H
 
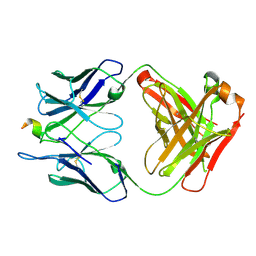 | |
1I8C
 
 | | SOLUTION STRUCTURE OF THE WATER-SOLUBLE FRAGMENT OF RAT HEPATIC APOCYTOCHROME B5 | | Descriptor: | CYTOCHROME B5 | | Authors: | Falzone, C.J, Wang, Y, Vu, B.C, Scott, N.L, Bhattacharya, S, Lecomte, J.T. | | Deposit date: | 2001-03-13 | | Release date: | 2001-05-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic perturbations induced by heme binding in cytochrome b5.
Biochemistry, 40, 2001
|
|
4BT0
 
 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR | | Authors: | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | Deposit date: | 2013-06-12 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8KER
 
 | |
8KDT
 
 | |
8KDS
 
 | |
