8B00
 
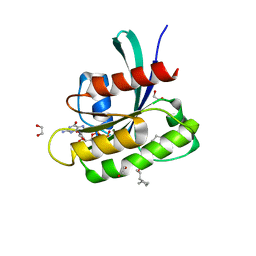 | | KRAS-G13D in complex with BI-2865 | | Descriptor: | (4S)-2-azanyl-4-methyl-4-[3-[4-[(1S)-1-[(2S)-1-methylpyrrolidin-1-ium-2-yl]ethoxy]pyrimidin-2-yl]-1,2,4-oxadiazol-5-yl]-6,7-dihydro-5H-1-benzothiophene-3-carbonitrile, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Boettcher, J, Herdeis, L. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Pan-KRAS inhibitor disables oncogenic signalling and tumour growth.
Nature, 619, 2023
|
|
8AZZ
 
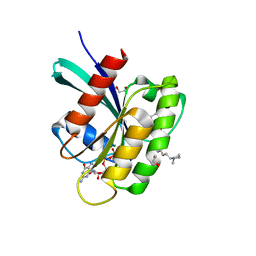 | | KRAS-G12V in complex with BI-2865 | | Descriptor: | (4S)-2-azanyl-4-methyl-4-[3-[4-[(1S)-1-[(2S)-1-methylpyrrolidin-1-ium-2-yl]ethoxy]pyrimidin-2-yl]-1,2,4-oxadiazol-5-yl]-6,7-dihydro-5H-1-benzothiophene-3-carbonitrile, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Boettcher, J, Herdeis, L. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Pan-KRAS inhibitor disables oncogenic signalling and tumour growth.
Nature, 619, 2023
|
|
7TZO
 
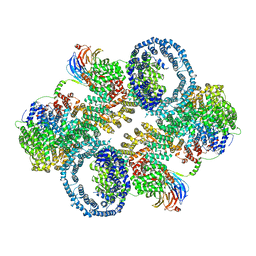 | | The apo structure of human mTORC2 complex | | Descriptor: | Rapamycin-insensitive companion of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex 2 subunit MAPKAP1, ... | | Authors: | Yu, Z, Chen, J, Pearce, D. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Interactions between mTORC2 core subunits Rictor and mSin1 dictate selective and context-dependent phosphorylation of substrate kinases SGK1 and Akt.
J.Biol.Chem., 298, 2022
|
|
7PUK
 
 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis in complex with Man5 product | | Descriptor: | Beta-N-acetylhexosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
7PUL
 
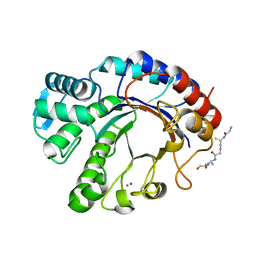 | | Crystal structure of Endoglycosidase E GH20 domain from Enterococcus faecalis | | Descriptor: | Beta-N-acetylhexosaminidase, CALCIUM ION, GLY-ALA-GLY-ALA-ALA, ... | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
7PUJ
 
 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis | | Descriptor: | Beta-N-acetylhexosaminidase, CHLORIDE ION, ZINC ION | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
5FED
 
 | | EGFR kinase domain in complex with a covalent aminobenzimidazole inhibitor. | | Descriptor: | Epidermal growth factor receptor, ~{N}-[7-methyl-1-[(3~{R})-1-propanoylazepan-3-yl]benzimidazol-2-yl]-3-(trifluoromethyl)benzamide | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2015-12-16 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Discovery of (R,E)-N-(7-Chloro-1-(1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl)-1H-benzo[d]imidazol-2-yl)-2-methylisonicotinamide (EGF816), a Novel, Potent, and WT Sparing Covalent Inhibitor of Oncogenic (L858R, ex19del) and Resistant (T790M) EGFR Mutants for the Treatment of EGFR Mutant Non-Small-Cell Lung Cancers.
J.Med.Chem., 59, 2016
|
|
6NBI
 
 | | Cryo-EM structure of parathyroid hormone receptor type 1 in complex with a long-acting parathyroid hormone analog and G protein | | Descriptor: | CHOLESTEROL, Gs protein alpha subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, L.-H, Ma, S, Sutkeviciute, I, Shen, D.-D, Zhou, X.E, de Waal, P.P, Li, C.-Y, Kang, Y, Clark, L.J, Jean-Alphonse, F.G, White, A.D, Xiao, K, Yang, D, Jiang, Y, Watanabe, T, Gardella, T.J, Melcher, K, Wang, M.-W, Vilardaga, J.-P, Xu, H.E, Zhang, Y. | | Deposit date: | 2018-12-07 | | Release date: | 2019-04-17 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and dynamics of the active human parathyroid hormone receptor-1.
Science, 364, 2019
|
|
6NBH
 
 | | Cryo-EM structure of parathyroid hormone receptor type 1 in complex with a long-acting parathyroid hormone analog and G protein | | Descriptor: | CHOLESTEROL, Gs protein alpha subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, L.-H, Ma, S, Sutkeviciute, I, Shen, D.-D, Zhou, X.E, de Waal, P.P, Li, C.-Y, Kang, Y, Clark, L.J, Jean-Alphonse, F.G, White, A.D, Xiao, K, Yang, D, Jiang, Y, Watanabe, T, Gardella, T.J, Melcher, K, Wang, M.-W, Vilardaga, J.-P, Xu, H.E, Zhang, Y. | | Deposit date: | 2018-12-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and dynamics of the active human parathyroid hormone receptor-1.
Science, 364, 2019
|
|
2K3K
 
 | |
3UX9
 
 | | Structural insights into a human anti-IFN antibody exerting therapeutic potential for systemic lupus erythematosus | | Descriptor: | Interferon alpha-1/13, ScFv antibody | | Authors: | Ouyang, S, Zhao, L.X, Liang, W, Shaw, N, Liu, Z.-J, Liang, M.-F. | | Deposit date: | 2011-12-04 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into a human anti-IFN antibody exerting therapeutic potential for systemic lupus erythematosus
J.Mol.Med., 2012
|
|
2HC1
 
 | | Engineered catalytic domain of protein tyrosine phosphatase HPTPbeta. | | Descriptor: | ACETATE ION, CHLORIDE ION, Receptor-type tyrosine-protein phosphatase beta | | Authors: | Evdokimov, A.G, Pokross, M, Walter, R, Mekel, M. | | Deposit date: | 2006-06-14 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2HC2
 
 | | Engineered protein tyrosine phosphatase beta catalytic domain | | Descriptor: | MAGNESIUM ION, Receptor-type tyrosine-protein phosphatase beta, SODIUM ION | | Authors: | Evdokimov, A.G, Pokross, M, Walter, R, Mekel, M. | | Deposit date: | 2006-06-15 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3QAH
 
 | | Crystal structure of apo-form human MOF catalytic domain | | Descriptor: | Probable histone acetyltransferase MYST1, ZINC ION | | Authors: | Sun, B, Tang, Q, Zhong, C, Ding, J. | | Deposit date: | 2011-01-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of the histone acetyltransferase activity of hMOF via autoacetylation of Lys274
Cell Res., 21, 2011
|
|
3K66
 
 | | X-ray crystal structure of the E2 domain of C. elegans APL-1 | | Descriptor: | Beta-amyloid-like protein | | Authors: | Hoopes, J.T, Ha, Y. | | Deposit date: | 2009-10-08 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of the E2 domain of APL-1, a Caenorhabditis elegans homolog of human amyloid precursor protein, and its heparin binding site
J.Biol.Chem., 285, 2010
|
|
3FSV
 
 | |
3K6B
 
 | | X-ray crystal structure of the E2 domain of APL-1 from C. elegans, in complex with sucrose octasulfate (SOS) | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Beta-amyloid-like protein | | Authors: | Hoopes, J.T, Ha, Y. | | Deposit date: | 2009-10-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of the E2 domain of APL-1, a Caenorhabditis elegans homolog of human amyloid precursor protein, and its heparin binding site
J.Biol.Chem., 285, 2010
|
|
3FSA
 
 | |
3FS9
 
 | |
3FT0
 
 | |
7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
3FSW
 
 | |
3FSZ
 
 | |
3K9X
 
 | | X-ray crystal structure of human fxa in complex with (S)-N-((2-METHYLBENZOFURAN-5-YLAMINO)(2-OXO-1-(2-OXO-2- (PYRROLIDIN-1-YL)ETHYL)AZEPAN-3- YLAMINO)METHYLENE)NICOTINAMIDE | | Descriptor: | CALCIUM ION, GLYCEROL, N-{N'-(2-methyl-1-benzofuran-5-yl)-N-[(3S)-2-oxo-1-(2-oxo-2-pyrrolidin-1-ylethyl)azepan-3-yl]carbamimidoyl}pyridine-3-carboxamide, ... | | Authors: | Klei, H.E, Kish, K, Ghosh, K, Rushith, A. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Aroylguanidine-based factor Xa inhibitors: the discovery of BMS-344577
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4HVB
 
 | | Catalytic unit of PI3Kg in complex with PI3K/mTOR dual inhibitor PF-04979064 | | Descriptor: | 1-{1-[(2S)-2-hydroxypropanoyl]piperidin-4-yl}-3-methyl-8-(6-methylpyridin-3-yl)-1,3-dihydro-2H-imidazo[4,5-c][1,5]naphthyridin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Knighton, D.R, Cheng, H. | | Deposit date: | 2012-11-05 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of the Highly Potent PI3K/mTOR Dual Inhibitor PF-04979064 through Structure-Based Drug Design.
ACS Med Chem Lett, 4, 2013
|
|
