7QWV
 
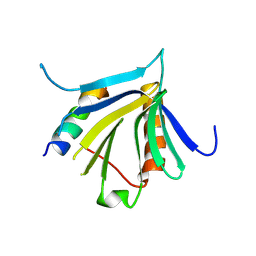 | | Crystal structure of the REC114-TOPOVIBL complex. | | Descriptor: | Meiotic recombination protein REC114, Type 2 DNA topoisomerase 6 subunit B-like | | Authors: | Juarez-Martinez, A.B, Robert, T, de Massy, B, Kadlec, J. | | Deposit date: | 2022-01-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | TOPOVIBL-REC114 interaction regulates meiotic DNA double-strand breaks.
Nat Commun, 13, 2022
|
|
8QS5
 
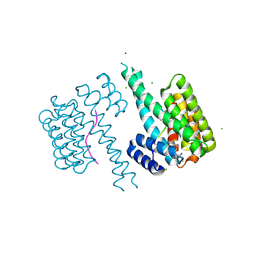 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 21 (1075354) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[3-fluoranyl-4-(trifluoromethyl)phenyl]sulfonylpiperidin-4-yl]methyl]ethanamide, C-RAF peptide pS259, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
8QSA
 
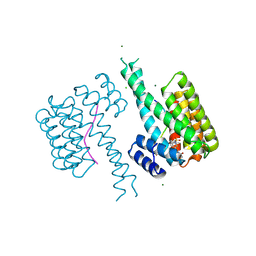 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 86 (1084384) | | Descriptor: | 1-[(5~{R})-2-(4-bromanyl-3-fluoranyl-phenyl)sulfonyl-2,7-diazaspiro[4.4]nonan-7-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, C-RAF peptide, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
8QSF
 
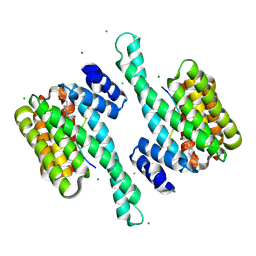 | | Ternary structure of 14-3-3s, BRAF phosphopeptide (pS365) and compound 22 (1083853). | | Descriptor: | 14-3-3 protein sigma, BRAF peptide pS365, CHLORIDE ION, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
8QS8
 
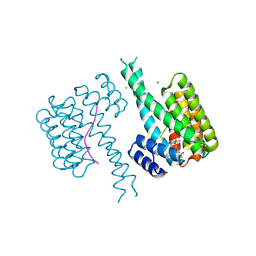 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 78 (1084378) | | Descriptor: | 1-[8-(4-bromophenyl)sulfonyl-5-oxa-2,8-diazaspiro[3.5]nonan-2-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, C-RAF peptide, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
8QS9
 
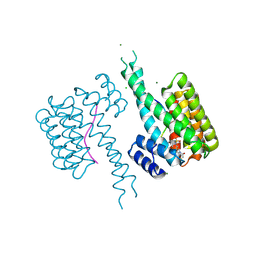 | | Ternary structure of 14-3-3s, C-RAF phosphopeptide (pS259) and compound 83 (1084383) | | Descriptor: | 1-[8-(4-bromanyl-3-fluoranyl-phenyl)sulfonyl-2,8-diazaspiro[3.5]nonan-2-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, C-RAF peptide pS259, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
8QSG
 
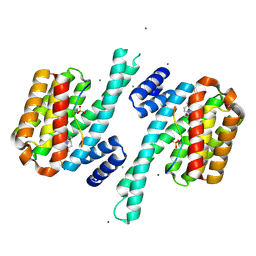 | | Ternary structure of 14-3-3s, BRAF phosphopeptide (pS365) and compound 86 (1124384). | | Descriptor: | 1-[(5~{R})-2-(4-bromanyl-3-fluoranyl-phenyl)sulfonyl-2,7-diazaspiro[4.4]nonan-7-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, BRAF peptide pS365, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
6T7U
 
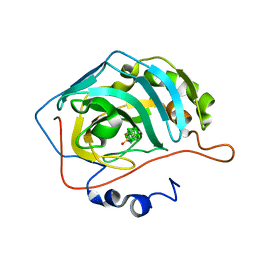 | | Carborane inhibitor of Carbonic Anhydrase IX | | Descriptor: | Carbonic anhydrase 2, Carborane inhibitor, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Kugler, M, Gruner, B. | | Deposit date: | 2019-10-23 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Sulfonamido carboranes as highly selective inhibitors of cancer-specific carbonic anhydrase IX.
Eur.J.Med.Chem., 200, 2020
|
|
8QSC
 
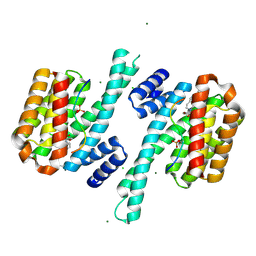 | | Ternary structure of 14-3-3s, ARAF phosphopeptide (pS214) and compound 22 (1083853). | | Descriptor: | 14-3-3 protein sigma, ARAF peptide pS214, CHLORIDE ION, ... | | Authors: | Konstantinidou, M, Vickery, H, Pennings, M.A.M, Virta, J, Visser, E.J, Oetelaar, M.C.M, Overmans, M, Neitz, J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule stabilization of the 14-3-3sigma/CRAF complex inhibits the MAPK pathway
To Be Published
|
|
1KQQ
 
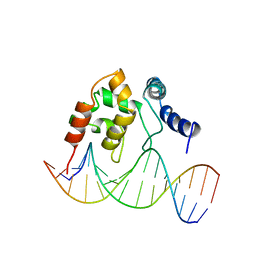 | | Solution Structure of the Dead ringer ARID-DNA Complex | | Descriptor: | 5'-D(*CP*CP*AP*CP*AP*TP*CP*AP*AP*TP*AP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*TP*AP*TP*TP*GP*AP*TP*GP*TP*GP*G)-3', DEAD RINGER PROTEIN | | Authors: | Iwahara, J, Iwahara, M, Daughdrill, G.W, Ford, J, Clubb, R.T. | | Deposit date: | 2002-01-07 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA.
EMBO J., 21, 2002
|
|
4LL4
 
 | | The structure of the TRX and TXNIP complex | | Descriptor: | Thioredoxin, Thioredoxin-interacting protein | | Authors: | Hwang, J, Kim, M.H. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein
Nat Commun, 5, 2014
|
|
6T9Z
 
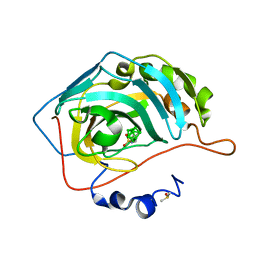 | | Nidocarborane inhibitor of Carbonic Anhydrase IX | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, Nidocarborane, ... | | Authors: | Brynda, J, Rezacova, P, Kugler, M, Gruner, B. | | Deposit date: | 2019-10-29 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Sulfonamido carboranes as highly selective inhibitors of cancer-specific carbonic anhydrase IX.
Eur.J.Med.Chem., 200, 2020
|
|
5JFW
 
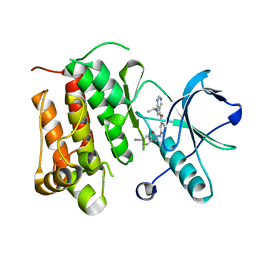 | | Crystal structure of TrkA in complex with PF-05247452 | | Descriptor: | 2-(4-cyanophenyl)-N-{5-[7-(propan-2-yl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonyl]pyridin-3-yl}acetamide, High affinity nerve growth factor receptor | | Authors: | Jayasankar, J, Kurumbail, R, Skerratt, S, Brown, D. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The Discovery of a Potent, Selective, and Peripherally Restricted Pan-Trk Inhibitor (PF-06273340) for the Treatment of Pain.
J. Med. Chem., 59, 2016
|
|
8RGW
 
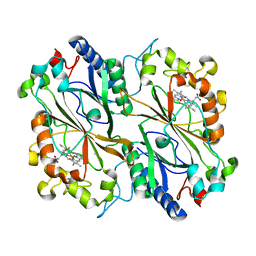 | | Serial synchrotron in plate room temperature structure of Dye Type Peroxidase Aa, 12 drops merged | | Descriptor: | Deferrochelatase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Thompson, A.J, Hough, M.A, Williams, L.J, Worrall, J.A.R, Sanchez-Weatherby, J, Mikolajek, H, Sandy, J. | | Deposit date: | 2023-12-14 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Efficient in situ screening of and data collection from microcrystals in crystallization plates.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RGY
 
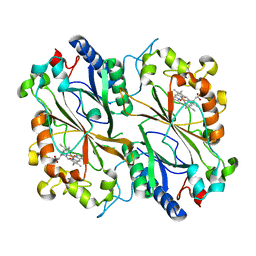 | | Serial synchrotron in plate room temperature structure of Dye Type Peroxidase Aa, 8 drops merged | | Descriptor: | Deferrochelatase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Thompson, A.J, Hough, M.A, Williams, L.J, Worrall, J.A.R, Sanchez-Weatherby, J, Mikolajek, H, Sandy, J. | | Deposit date: | 2023-12-14 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Efficient in situ screening of and data collection from microcrystals in crystallization plates.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RGE
 
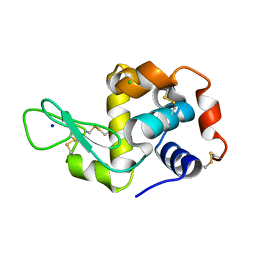 | | Serial synchrotron in plate room temperature structure of Lysozyme. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Thompson, A.J, Hough, M.A, Sanchez-Weatherby, J, Williams, L.J, Sandy, J, Worrall, J.A.R. | | Deposit date: | 2023-12-13 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Efficient in situ screening of and data collection from microcrystals in crystallization plates.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RGS
 
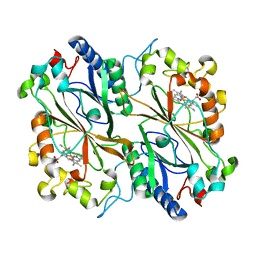 | | Serial synchrotron in plate room temperature structure of Dye Type Peroxidase Aa | | Descriptor: | Deferrochelatase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Thompson, A.J, Hough, M.A, Williams, L.J, Worrall, J.A.R, Sanchez-Weatherby, J, Mikolajek, H, Sandy, J. | | Deposit date: | 2023-12-14 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Efficient in situ screening of and data collection from microcrystals in crystallization plates.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4LCW
 
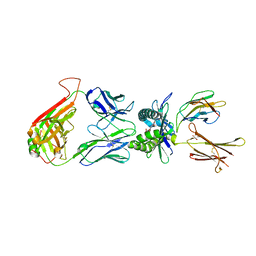 | | The structure of human MAIT TCR in complex with MR1-K43A-RL-6-Me-7OH | | Descriptor: | 1-deoxy-1-(7-hydroxy-6-methyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Patel, O, Rossjohn, J. | | Deposit date: | 2013-06-24 | | Release date: | 2013-10-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells.
J.Exp.Med., 210, 2013
|
|
5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5JOW
 
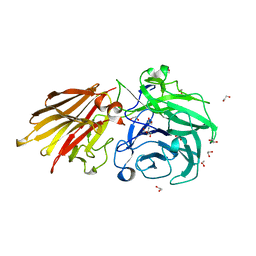 | | Bacteroides ovatus Xyloglucan PUL GH43A | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-reducing end alpha-L-arabinofuranosidase BoGH43A | | Authors: | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
3SKO
 
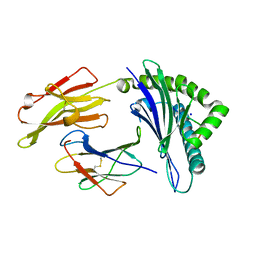 | | Crystal structure of the HLA-B8-A66-FLR, mutant A66 of the HLA B8 | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 3, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
5JP0
 
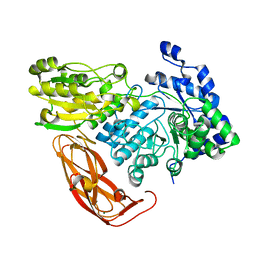 | | Bacteroides ovatus Xyloglucan PUL GH3B with bound glucose | | Descriptor: | Beta-glucosidase BoGH3B, MAGNESIUM ION, beta-D-glucopyranose | | Authors: | Hemsworth, G.R, Thompson, A.J, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
5JOU
 
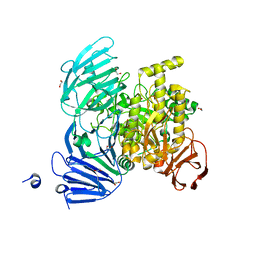 | | Bacteroides ovatus Xyloglucan PUL GH31 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase BoGH31A, NICKEL (II) ION | | Authors: | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
5JQM
 
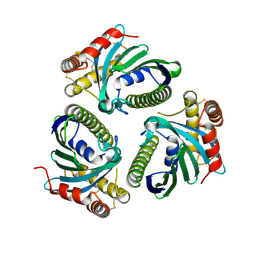 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 Void of Bound Phospholipid from Saccharomyces Cerevisiae at 1.5 Angstroms Resolution | | Descriptor: | Protein UPS1, mitochondrial,Mitochondrial distribution and morphology protein 35 | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2016-05-05 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1
Commun Biol, 2020
|
|
3SKM
 
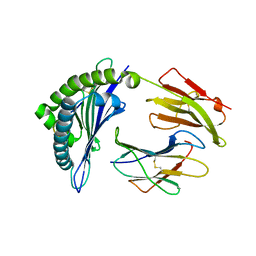 | | Crystal structure of the HLA-B8FLRGRAYVL, mutant G8V of the FLR peptide | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 3, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
