5GX9
 
 | | PYP mutant - E46Q | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Yonezawa, K. | | Deposit date: | 2016-09-16 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | NEUTRON DIFFRACTION (1.493 Å) | | Cite: | Neutron crystallography of photoactive yellow protein reveals unusual protonation state of Arg52 in the crystal
Sci Rep, 7, 2017
|
|
8I5F
 
 | | Crystal structure of the DHR-2 domain of DOCK10 in complex with Cdc42 (T17N mutant) | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 10 | | Authors: | Kukimoto-Niino, M, Mishima-Tsumagari, C, Fukui, Y, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2023-01-25 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the dual GTPase specificity of the DOCK10 guanine nucleotide exchange factor.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
1UK1
 
 | |
1VCV
 
 | |
1MPG
 
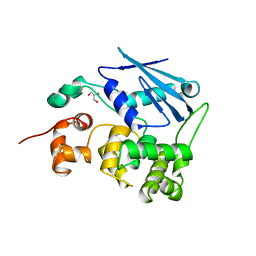 | | 3-METHYLADENINE DNA GLYCOSYLASE II FROM ESCHERICHIA COLI | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE II, GLYCEROL | | Authors: | Labahn, J, Schaerer, O.D, Long, A, Ezaz-Nikpay, K, Verdine, G.L, Ellenberger, T.E. | | Deposit date: | 1997-10-28 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the excision repair of alkylation-damaged DNA.
Cell(Cambridge,Mass.), 86, 1996
|
|
5GSE
 
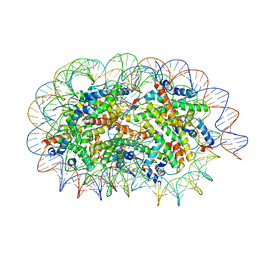 | | Crystal structure of unusual nucleosome | | Descriptor: | DNA (250-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kato, D, Osakabe, A, Arimura, Y, Park, S.Y, Kurumizaka, H. | | Deposit date: | 2016-08-16 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of the overlapping dinucleosome composed of hexasome and octasome
Science, 356, 2017
|
|
1GEQ
 
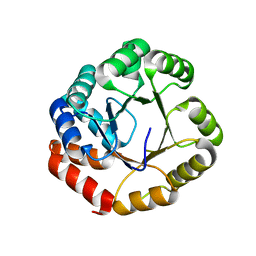 | |
4L35
 
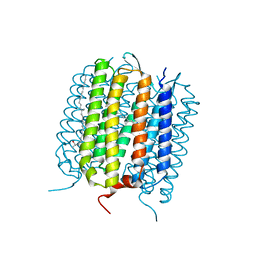 | |
1GEJ
 
 | | STRUCTURAL CHARACTERIZATION OF N-BUTYL-ISOCYANIDE COMPLEXES OF CYTOCHROMES P450NOR AND P450CAM | | Descriptor: | CYTOCHROME P450 55A1, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | lee, D.-S, Park, S.-Y, Yamane, K, Shiro, Y. | | Deposit date: | 2000-11-13 | | Release date: | 2000-12-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of n-butyl-isocyanide complexes of cytochromes P450nor and P450cam.
Biochemistry, 40, 2001
|
|
3WXQ
 
 | | Serial femtosecond X-ray structure of human fatty acid-binding protein type-3 (FABP3) in complex with stearic acid (C18:0) determined using X-ray free-electron laser at SACLA | | Descriptor: | Fatty acid-binding protein, heart, STEARIC ACID | | Authors: | Mizohata, E, Suzuki, M, Kakinouchi, K, Sugiyama, S, Murata, M, Sugahara, M, Nango, E, Tanaka, T, Tanaka, R, Tono, K, Song, C, Hatsui, T, Joti, Y, Yabashi, M, Iwata, S. | | Deposit date: | 2014-08-04 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
3FX5
 
 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by High Resolution X-ray Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, GLYCEROL, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
1GEI
 
 | | STRUCTURAL CHARACTERIZATION OF N-BUTYL-ISOCYANIDE COMPLEXES OF CYTOCHROMES P450NOR AND P450CAM | | Descriptor: | CYTOCHROME P450 55A1, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, D.-S, Park, S.-Y, Yamane, K, Shiro, Y. | | Deposit date: | 2000-11-13 | | Release date: | 2000-11-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of n-butyl-isocyanide complexes of cytochromes P450nor and P450cam.
Biochemistry, 40, 2001
|
|
5B0W
 
 | |
6A3J
 
 | | Levoglucosan dehydrogenase, complex with NADH and L-sorbose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative dehydrogenase, ... | | Authors: | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
3WUM
 
 | |
5WL0
 
 | | Co-crystal structure of Influenza A H3N2 PB2 (241-741) bound to VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, Polymerase basic protein 2 | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for therapeutic inhibition of influenza A polymerase PB2 subunit.
Sci Rep, 7, 2017
|
|
5WZY
 
 | | Crystal structure of the P2X4 receptor from zebrafish in the presence of CTP at 2.8 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Kasuya, G, Hattori, M, Nureki, O. | | Deposit date: | 2017-01-19 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural insights into the nucleotide base specificity of P2X receptors
Sci Rep, 7, 2017
|
|
3WUL
 
 | |
3WXS
 
 | | Thaumatin structure determined by SPring-8 Angstrom Compact free electron Laser (SACLA) | | Descriptor: | L(+)-TARTARIC ACID, thaumatin I | | Authors: | Masuda, T, Nango, E, Sugahara, M, Mizohata, E, Tanaka, T, Tanaka, R, Suzuki, M, Mikami, B, Iwata, S. | | Deposit date: | 2014-08-07 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
3WXT
 
 | |
3WXU
 
 | |
3HGP
 
 | | Structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180 determined by high resolution crystallography | | Descriptor: | 4-[[(2S)-3-methyl-1-oxo-1-[(2S)-2-[[(3S)-1,1,1-trifluoro-4-methyl-2-oxo-pentan-3-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamoyl]benzoic acid, CALCIUM ION, Elastase-1, ... | | Authors: | Tamada, T, Kinoshita, T, Kuroki, R, Tada, T. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Combined High-Resolution Neutron and X-ray Analysis of Inhibited Elastase Confirms the Active-Site Oxyanion Hole but Rules against a Low-Barrier Hydrogen Bond
J.Am.Chem.Soc., 131, 2009
|
|
1BAK
 
 | |
5W2P
 
 | | Crystal structure of Mycobacterium tuberculosis KasA in complex with 6U5 | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 1, GLYCEROL, ... | | Authors: | Capodagli, G.C, Neiditch, M.B. | | Deposit date: | 2017-06-06 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synergistic Lethality of a Binary Inhibitor of Mycobacterium tuberculosis KasA.
MBio, 9, 2018
|
|
6A3I
 
 | | Levoglucosan dehydrogenase, complex with NADH and levoglucosan | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Levoglucosan, Putative dehydrogenase | | Authors: | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
