6X1C
 
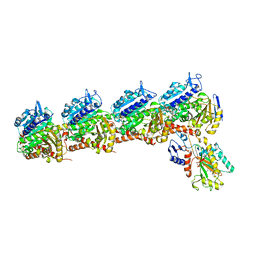 | | Tubulin-RB3_SLD-TTL in complex with compound 5j | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chlorofuro[3,2-d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
6X1F
 
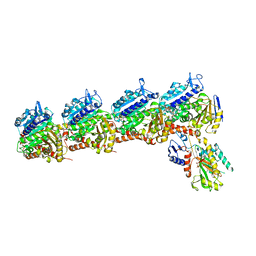 | | Tubulin-RB3_SLD-TTL in complex with compound 5m | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-methoxy-4-(2-methyl-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl)-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
6X1E
 
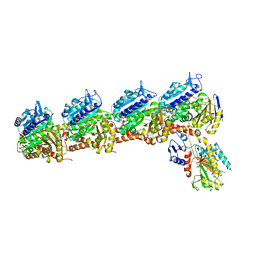 | | Tubulin-RB3_SLD-TTL in complex with compound 5l | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chloro-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
6LAS
 
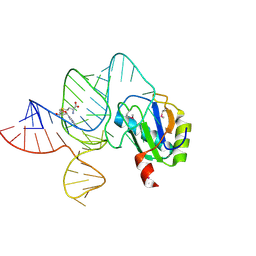 | | the wildtype SAM-VI riboswitch bound to SAM | | Descriptor: | RNA (55-MER), S-ADENOSYLMETHIONINE, U1 small nuclear ribonucleoprotein A | | Authors: | Ren, A, Sun, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
6LIS
 
 | | ASFV dUTPase in complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6LJ3
 
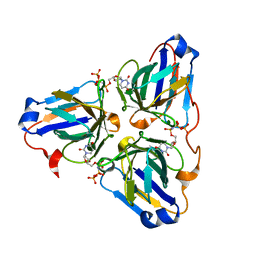 | |
6LJO
 
 | | African swine fever virus dUTPase | | Descriptor: | E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
3A1L
 
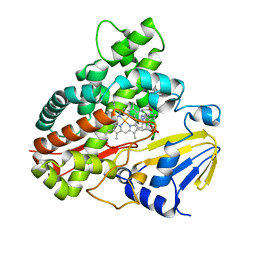 | | Crystal Structure of 11,11'-Dichlorochromopyrrolic Acid Bound Cytochrome P450 StaP (CYP245A1) | | Descriptor: | 3,4-bis(7-chloro-1H-indol-3-yl)-1H-pyrrole-2,5-dicarboxylic acid, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Makino, M, Sugimoto, H, Shiro, Y, Asamizu, S, Onaka, H, Nagano, S. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Theoretical and experimental studies of the conversion of chromopyrrolic acid to an antitumor derivative by cytochrome P450 StaP: the catalytic role of water molecules
J.Am.Chem.Soc., 131, 2009
|
|
7SSA
 
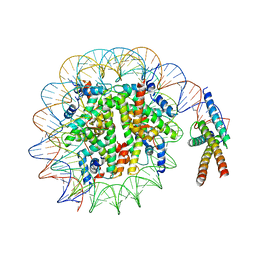 | | Cryo-EM structure of pioneer factor Cbf1 bound to the nucleosome | | Descriptor: | Centromere-binding protein 1, DNA (137-MER), Histone H2A.1, ... | | Authors: | Eek, P, Tan, S. | | Deposit date: | 2021-11-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Basic helix-loop-helix pioneer factors interact with the histone octamer to invade nucleosomes and generate nucleosome-depleted regions.
Mol.Cell, 83, 2023
|
|
7TL3
 
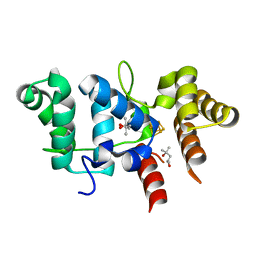 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 5YF431 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, IRON/SULFUR CLUSTER | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.066 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
7TL4
 
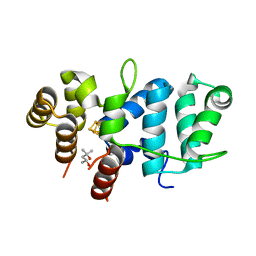 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 6YF | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, IRON/SULFUR CLUSTER | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
7TL2
 
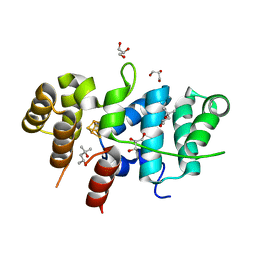 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 5YF412 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, GLYCEROL, ... | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
6AH3
 
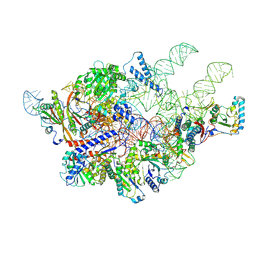 | | Cryo-EM structure of yeast Ribonuclease P with pre-tRNA substrate | | Descriptor: | MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
5EG3
 
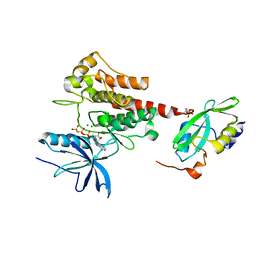 | | Crystal Structure of the Activated FGF Receptor 2 (FGFR2) Kinase Domain in complex with the cSH2 domain of Phospholipase C gamma (PLCgamma) | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1, Fibroblast growth factor receptor 2, MAGNESIUM ION, ... | | Authors: | Huang, Z, Li, X, Mohammadi, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Two FGF Receptor Kinase Molecules Act in Concert to Recruit and Transphosphorylate Phospholipase C gamma.
Mol.Cell, 61, 2016
|
|
9JS4
 
 | | Cryo-EM structure of neutralizing antibody 8G3 in complex with BA.1 RBD | | Descriptor: | Heavy chain of 8G3, Light chain of 8G3, Spike glycoprotein | | Authors: | Li, J, Li, H. | | Deposit date: | 2024-09-30 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Rapid restoration of potent neutralization activity against the latest Omicron variant JN.1 via AI rational design and antibody engineering.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
1JR7
 
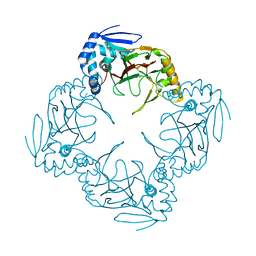 | |
5GJT
 
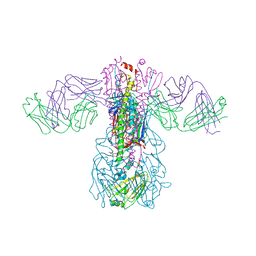 | | Crystal structure of H1 hemagglutinin from A/Washington/05/2011 in complex with a neutralizing antibody 3E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, heavy chain of human neutralizing antibody 3E1, ... | | Authors: | Wang, W, Zhang, T, Ding, J. | | Deposit date: | 2016-07-01 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Human antibody 3E1 targets the HA stem region of H1N1 and H5N6 influenza A viruses
Nat Commun, 7, 2016
|
|
7V2Y
 
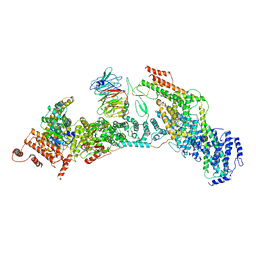 | | cryo-EM structure of yeast THO complex with Sub2 | | Descriptor: | ATP-dependent RNA helicase SUB2, Protein TEX1, THO complex subunit 2, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
7V2W
 
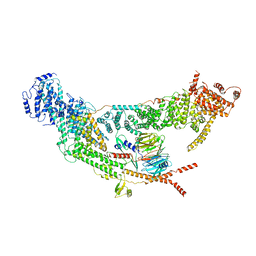 | | protomer structure from the dimer of yeast THO complex | | Descriptor: | Protein TEX1, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
5GSX
 
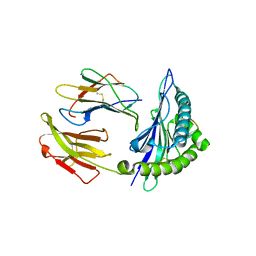 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-2 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
6AGB
 
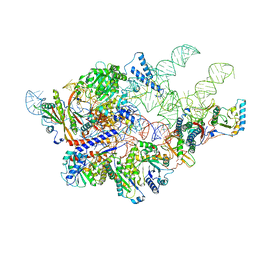 | | Cryo-EM structure of yeast Ribonuclease P | | Descriptor: | RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, Ribonuclease P protein subunit RPR2, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
8YLE
 
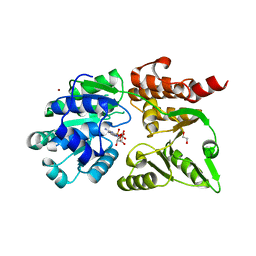 | | Crystal structure of Werner syndrome helicase complexed with AMP-PCP | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Yang, Y, Fu, L, Sun, X, Cheng, H, Chen, R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of werner syndrome helicase complexed with AMP-PCP at 1.86 Angstroms resolution.
To Be Published
|
|
6Q1P
 
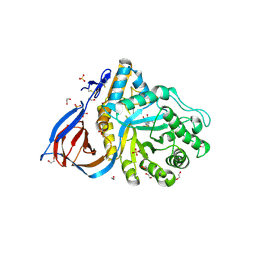 | | Glucocerebrosidase in complex with pharmacological chaperone norIMX8 | | Descriptor: | (2S,3S,4S,5R)-2-{[(4-methylpentyl)sulfonyl]methyl}piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vickers, C, Withers, S.G, Boraston, A.B. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Pharmacological chaperones for GCase that switch conformation with pH enhance enzyme levels in Gaucher animal models.
To Be Published
|
|
6Q1N
 
 | | Glucocerebrosidase in complex with pharmacological chaperone IMX8 | | Descriptor: | (2R,3S,4S,5R)-2-[2-(methylsulfanyl)ethyl]piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vickers, C, Withers, S.G, Boraston, A.B. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.526 Å) | | Cite: | Pharmacological chaperones for GCase that switch conformation with pH enhance enzyme levels in Gaucher animal models.
To Be Published
|
|
5WYS
 
 | | luciferase with inhibitor 3i | | Descriptor: | 5-[(3R)-3-(4-boranylphenyl)-3-oxidanyl-propyl]-2-oxidanyl-benzoic acid, Luciferin 4-monooxygenase | | Authors: | Gu, L, Su, J, Wang, F. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Inhibiting Firefly Bioluminescence by Chalcones
Anal. Chem., 89, 2017
|
|
