7NII
 
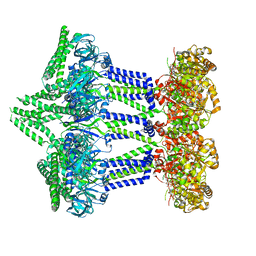 | | Wzc-K540M MgADP C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
7NI2
 
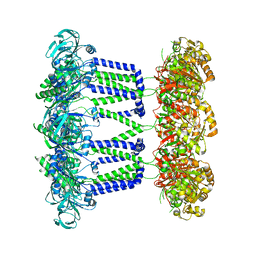 | | Wzc-K540M-4YE C8 | | Descriptor: | Tyrosine-protein kinase | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
7NIB
 
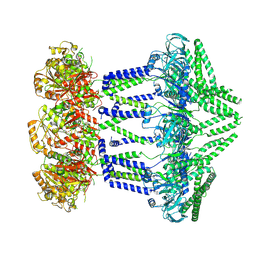 | | Wzc-K540M-4YE C1 | | Descriptor: | Tyrosine-protein kinase | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
7NG7
 
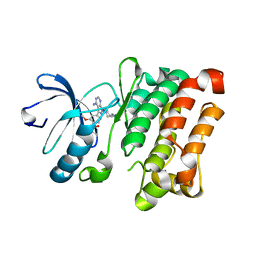 | | Src kinase bound to eCF506 trapped in inactive conformation | | Descriptor: | 1,2-ETHANEDIOL, Proto-oncogene tyrosine-protein kinase Src, tert-butyl (4-(4-amino-1-(2-(4-(dimethylamino)piperidin-1-yl)ethyl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl)-2-methoxyphenyl)carbamate | | Authors: | Lietha, D, Unciti-Broceta, A. | | Deposit date: | 2021-02-08 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Conformation Selective Mode of Inhibiting SRC Improves Drug Efficacy and Tolerability.
Cancer Res., 81, 2021
|
|
2M6U
 
 | | NMR Structure of CbpAN from Streptococcus pneumoniae | | Descriptor: | Choline binding protein A | | Authors: | Liu, A, Yan, H, Achila, D, Martinez-Hackert, E, Li, Y, Banerjee, R. | | Deposit date: | 2013-04-10 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of host specificity of complement Factor H recruitment by Streptococcus pneumoniae.
Biochem.J., 465, 2015
|
|
7N4I
 
 | |
7N4J
 
 | |
7N12
 
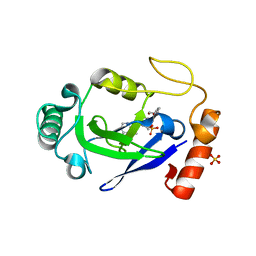 | | Crystal structure of the M. abscessus LeuRS editing domain in complex with epetraborole-AMP adduct | | Descriptor: | Leucine--tRNA ligase, SULFATE ION, [(1S,5R,6R,7'S,8R)-7'-(aminomethyl)-6-(6-aminopurin-9-yl)-2'-(3-oxidanylpropoxy)spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,9'-8-oxa-9-boranuidabicyclo[4.3.0]nona-1(6),2,4-triene]-8-yl]methyl dihydrogen phosphate | | Authors: | Kalthoff, E, Schmeing, M. | | Deposit date: | 2021-05-26 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Efficacy of epetraborole against Mycobacterium abscessus is increased with norvaline.
Plos Pathog., 17, 2021
|
|
7N11
 
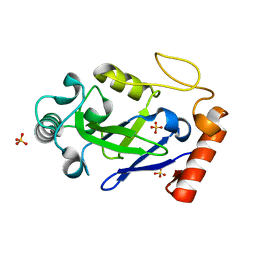 | |
7NL4
 
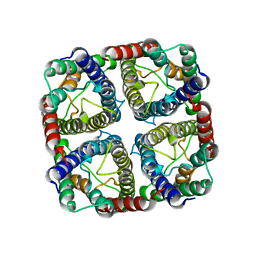 | | OsNIP2;1 silicon transporter from rice | | Descriptor: | Aquaporin NIP2-1, CADMIUM ION | | Authors: | van den Berg, B. | | Deposit date: | 2021-02-22 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Silicic Acid Uptake by Higher Plants.
J.Mol.Biol., 433, 2021
|
|
7N4M
 
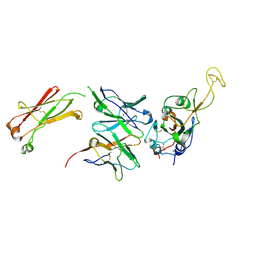 | |
7MXE
 
 | | Ab1245 Fab in complex with BG505 SOSIP.664 and 8ANC195 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 8ANC195 G52K5 Fab heavy chain, ... | | Authors: | Abernathy, M.E, Bjorkman, P.J. | | Deposit date: | 2021-05-19 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Antibody elicited by HIV-1 immunogen vaccination in macaques displaces Env fusion peptide and destroys a neutralizing epitope.
Npj Vaccines, 6, 2021
|
|
4G8G
 
 | | Crystal Structure of C12C TCR-HA B2705-KK10 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-27 alpha chain, ... | | Authors: | Gras, S, Wilmann, P.G, Rossjohn, J. | | Deposit date: | 2012-07-23 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Molecular Basis for the Control of Preimmune Escape Variants by HIV-Specific CD8(+) T Cells.
Immunity, 38, 2013
|
|
7N4L
 
 | |
2MA4
 
 | | Solution NMR Structure of yahO protein from Salmonella typhimurium, Northeast Structural Genomics Consortium (NESG) Target StR106 | | Descriptor: | Putative periplasmic protein | | Authors: | Eletsky, A, Zhang, Q, Liu, G, Wang, H, Nwosu, C, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
7N9L
 
 | | KirBac3.1 C71S C262S | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Inward rectifier potassium channel Kirbac3.1, POTASSIUM ION, ... | | Authors: | Gulbis, J.M, Black, K.A. | | Deposit date: | 2021-06-18 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ion currents through Kir potassium channels are gated by anionic lipids.
Nat Commun, 13, 2022
|
|
3T9A
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP at pH 7.0 | | Descriptor: | CADMIUM ION, Inositol Pyrophosphate Kinase, MAGNESIUM ION, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
7N9K
 
 | | KirBac3.1 L124M mutant | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Inward rectifier potassium channel Kirbac3.1, N-OCTANE, ... | | Authors: | Black, T.A, Gulbis, J.M. | | Deposit date: | 2021-06-18 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Ion currents through Kir potassium channels are gated by anionic lipids.
Nat Commun, 13, 2022
|
|
4DVW
 
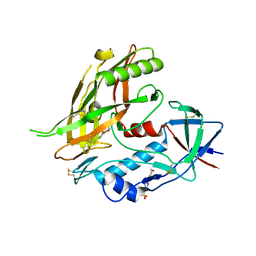 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with MAE-II-167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-(4-chloro-3-fluorophenyl)-N'-[(3aS,6aS)-hexahydrocyclopenta[c]pyrrol-3a(1H)-ylmethyl]ethanediamide, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2012-02-23 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of HIV-1 gp120 Envelope Glycoprotein in Complex with NBD Analogues That Target the CD4-Binding Site.
Plos One, 9, 2014
|
|
4DA5
 
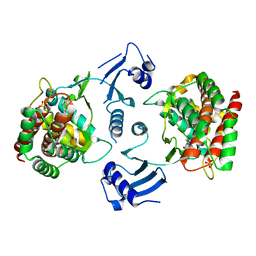 | | Choline Kinase alpha acts through a double-displacement kinetic mechanism involving enzyme isomerisation, as determined through enzyme and inhibitor kinetics and structural biology | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl[bis(5-chlorothiophen-2-yl)]methanol, Choline kinase alpha, SULFATE ION | | Authors: | Brown, K, Hudson, C, Charlton, P, Pollard, J. | | Deposit date: | 2012-01-12 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and mechanistic characterisation of Choline Kinase-alpha.
Biochim.Biophys.Acta, 1834, 2013
|
|
3T9D
 
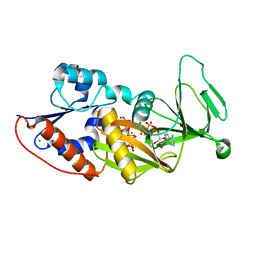 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP and 5-(PP)-IP5 (5-IP7) | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Inositol Pyrophosphate Kinase, MAGNESIUM ION, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3SSN
 
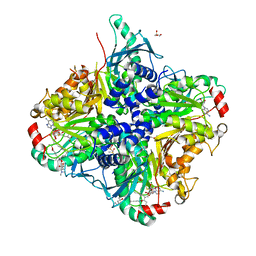 | | MycE Methyltransferase from the Mycinamycin Biosynthetic Pathway in Complex with Mg, SAH, and Mycinamycin VI | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | A new structural form in the SAM/metal-dependent o‑methyltransferase family: MycE from the mycinamicin biosynthetic pathway.
J.Mol.Biol., 413, 2011
|
|
3SSO
 
 | |
3T53
 
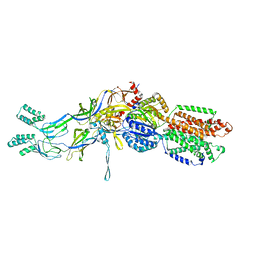 | | Crystal structures of the extrusion state of the CusBA adaptor-transporter complex | | Descriptor: | COPPER (II) ION, Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E.W. | | Deposit date: | 2011-07-26 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
2N71
 
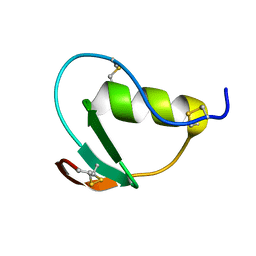 | | NMR structure of CmPI-II, a serin protease inhibitor isolated from mollusk Cenchitis muricatus | | Descriptor: | Protease inhibitor 2 | | Authors: | Cabrera-Munoz, A, Rojas, L, Alonso del Rivero Antigua, M, Pires, J. | | Deposit date: | 2015-09-01 | | Release date: | 2016-09-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of CmPI-II, a non-classical Kazal protease inhibitor: Understanding its conformational dynamics and subtilisin A inhibition.
J.Struct.Biol., 206, 2019
|
|
