6ADL
 
 | |
6ADS
 
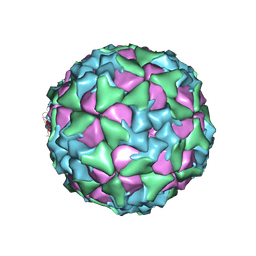 | | Structure of Seneca Valley Virus in acidic conditions | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Lou, Z.Y, Cao, L. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Seneca Valley virus attachment and uncoating mediated by its receptor anthrax toxin receptor 1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6ADT
 
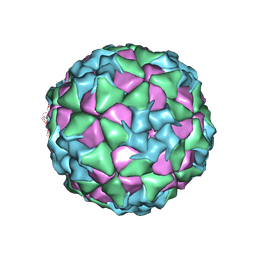 | | Structure of Seneca Valley Virus in neutral condition | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Lou, Z.Y, Cao, L. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Seneca Valley virus attachment and uncoating mediated by its receptor anthrax toxin receptor 1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5T1T
 
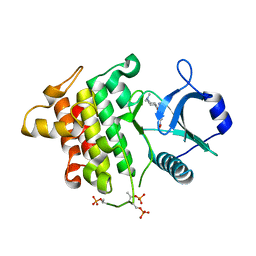 | | Irak4 kinase - compound 1 co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N},~{N}-dimethyl-4-(6-nitroquinazolin-4-yl)oxy-cyclohexan-1-amine | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Identification of quinazoline based inhibitors of IRAK4 for the treatment of inflammation.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T1S
 
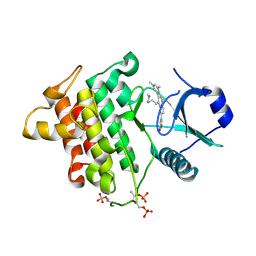 | | Irak4 kinase - compound 1 co-structure | | Descriptor: | 5-[3-(3,5-dimethylphenyl)-4-[4-(methylamino)butyl]quinolin-6-yl]pyridin-3-ol, Interleukin-1 receptor-associated kinase 4 | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of quinazoline based inhibitors of IRAK4 for the treatment of inflammation.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6ADM
 
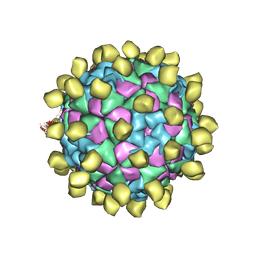 | |
5KJL
 
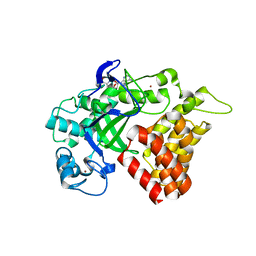 | | SMYD2 in complex with AZ378 | | Descriptor: | N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-20 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors.
J. Med. Chem., 59, 2016
|
|
4PHW
 
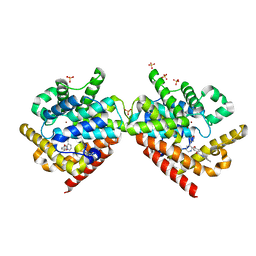 | |
6SFC
 
 | | EED in complex with a methyl-thiazole | | Descriptor: | CALCIUM ION, N-(1,3-benzodioxol-4-ylmethyl)-4-methyl-5-(1-methylpyrazol-3-yl)-1,3-thiazol-2-amine, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Read, J.A. | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rapid Identification of Novel Allosteric PRC2 Inhibitors.
Acs Chem.Biol., 14, 2019
|
|
8K6N
 
 | | Structural complex of neuropeptide Y receptor 2 | | Descriptor: | C-flanking peptide of NPY, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shen, S.Y, Zhao, C, Shao, Z.H. | | Deposit date: | 2023-07-25 | | Release date: | 2024-07-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of neuropeptide Y signaling through Y 1 and Y 2 receptors.
MedComm (2020), 5, 2024
|
|
8K6M
 
 | | Structural complex of neuropeptide Y receptor 1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | Authors: | Shen, S.Y, Zhao, C, Shao, Z.H. | | Deposit date: | 2023-07-25 | | Release date: | 2024-07-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of neuropeptide Y signaling through Y 1 and Y 2 receptors.
MedComm (2020), 5, 2024
|
|
8K6O
 
 | | Structural complex of neuropeptide Y receptor 1 | | Descriptor: | C-flanking peptide of NPY, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shen, S.Y, Zhao, C, Shao, Z.H. | | Deposit date: | 2023-07-25 | | Release date: | 2024-07-31 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of neuropeptide Y signaling through Y 1 and Y 2 receptors.
MedComm (2020), 5, 2024
|
|
6SFB
 
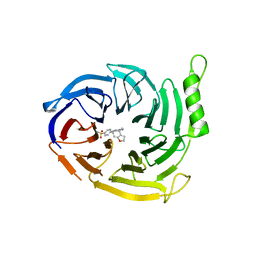 | | EED in complex with a triazolopyrimidine | | Descriptor: | GLYCEROL, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, Polycomb protein EED | | Authors: | Read, J.A. | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Rapid Identification of Novel Allosteric PRC2 Inhibitors.
Acs Chem.Biol., 14, 2019
|
|
4RZF
 
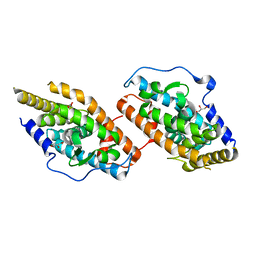 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, S441W mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
1BJ1
 
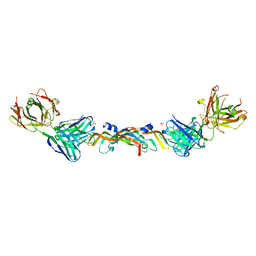 | | VASCULAR ENDOTHELIAL GROWTH FACTOR IN COMPLEX WITH A NEUTRALIZING ANTIBODY | | Descriptor: | Fab fragment, heavy chain, light chain, ... | | Authors: | Muller, Y.A, Christinger, H.W, De Vos, A.M. | | Deposit date: | 1998-06-30 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | VEGF and the Fab fragment of a humanized neutralizing antibody: crystal structure of the complex at 2.4 A resolution and mutational analysis of the interface.
Structure, 6, 1998
|
|
4RZG
 
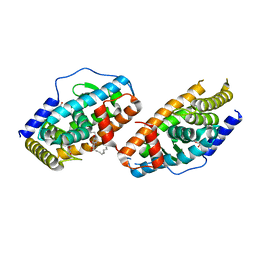 | | Crystal Structure Analysis of the DNPA-bounded NUR77 Ligand binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, pentyl (3,5-dihydroxy-2-nonanoylphenyl)acetate | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
1G1Z
 
 | | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus that Selectively Acts on Vertebrate Neuronal Na+ Channels, LEU12-PRO13 Cis isomer | | Descriptor: | CONOTOXIN EVIA | | Authors: | Volpon, L, Lamthanh, H, Le Gall, F, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-16 | | Release date: | 2000-11-01 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus That Selectively Acts on Vertebrate Neuronal Na+ Channels.
J.Biol.Chem., 279, 2004
|
|
1G1P
 
 | | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus that Selectively Acts on Vertebrate Neuronal Na+ Channels | | Descriptor: | CONOTOXIN EVIA | | Authors: | Volpon, L, Lamthanh, H, Barbier, J, Gilles, N, Molgo, J, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-13 | | Release date: | 2000-11-01 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus That Selectively Acts on Vertebrate Neuronal Na+ Channels.
J.Biol.Chem., 279, 2004
|
|
4BB4
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N-(2-methoxyethyl)-4-[(6-pyridin-4-ylquinazolin-2-yl)amino]benzamide | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Optimization of a Novel Series of Potent Mutant B-Raf V600E Selective Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
6LBR
 
 | | Crystal structure of yeast Cdc13 and ssDNA | | Descriptor: | KLLA0F20922p, Telomere single-strand DNA | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WQX
 
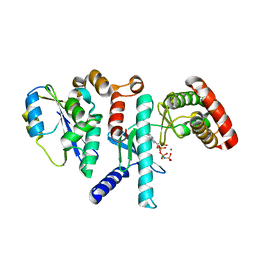 | | Human PRPK-TPRKB complex | | Descriptor: | EKC/KEOPS complex subunit TP53RK, EKC/KEOPS complex subunit TPRKB, MAGNESIUM ION, ... | | Authors: | Li, J, Ma, X.L, Banerjee, S, Dong, Z.G. | | Deposit date: | 2020-04-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structure of the human PRPK-TPRKB complex.
Commun Biol, 4, 2021
|
|
6LBT
 
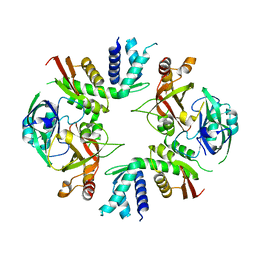 | | Crystal structure of yeast Cdc13 and Stn1 | | Descriptor: | GLYCEROL, KLLA0C11825p, KLLA0F20922p,KLLA0F20922p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LBU
 
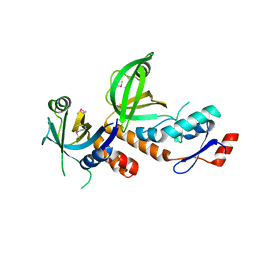 | | Crystal structure of yeast Stn1 and Ten1 | | Descriptor: | KLLA0C11825p, KLLA0E09417p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LAX
 
 | | the mutant SAM-VI riboswitch (U6C) bound to SAM | | Descriptor: | RNA (55-MER), S-ADENOSYLMETHIONINE, U1 small nuclear ribonucleoprotein A | | Authors: | Sun, A, Ren, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
6LAZ
 
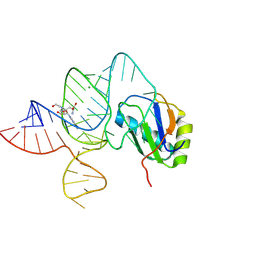 | | the wildtype SAM-VI riboswitch bound to a N-mustard SAM analog M1 | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(2-hydroxyethyl)amino]-2-azaniumyl-butanoate, MAGNESIUM ION, RNA (55-MER), ... | | Authors: | Ren, A, Sun, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
