4MSV
 
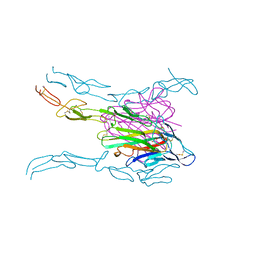 | | Crystal structure of FASL and DcR3 complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 6, ... | | Authors: | Liu, W, Ramagopal, U.A, Zhan, C, Bonanno, J.B, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-18 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Complex of Human FasL and Its Decoy Receptor DcR3.
Structure, 24, 2016
|
|
4N15
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Burkholderia ambifaria (BAM_6123), Target EFI-510059, with bound beta-D-glucuronate | | Descriptor: | MAGNESIUM ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N23
 
 | | Crystal structure of the GP2 Core Domain from the California Academy of Science Virus, monoclinic symmetry | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, GP2 Ectodomain | | Authors: | Malashkevich, V.N, Koellhoffer, J.F, Dai, Z, Toro, R, Lai, J.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of the Glycoprotein GP2 Core Domain from the CAS Virus, a Novel Arenavirus-Like Species.
J.Mol.Biol., 426, 2014
|
|
4N2Y
 
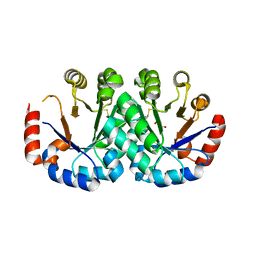 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Archaeoglobus fulgidus | | Descriptor: | GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-10-06 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Archaeoglobus fulgidus
To be Published
|
|
4MMW
 
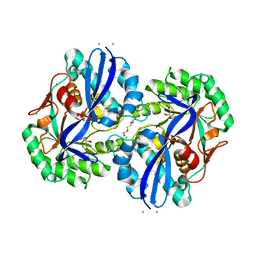 | | Crystal structure of D-glucarate dehydratase from Agrobacterium tumefaciens complexed with magnesium, L-Xylarohydroxamate and L-Lyxarohydroxamate | | Descriptor: | (2R,3S,4R)-2,3,4-TRIHYDROXY-5-(HYDROXYAMINO)-5-OXOPENTANOIC ACID, CHLORIDE ION, Isomerase/lactonizing enzyme, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-09-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal structure of D-glucarate dehydratase from Agrobacterium tumefaciens complexed with magnesium, L-Xylarohydroxamate and L-Lyxarohydroxamate
To be Published
|
|
4OLQ
 
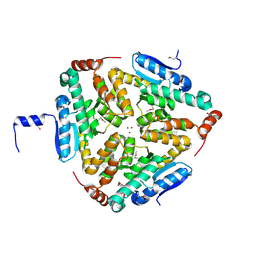 | | Crystal Structure of a Putative enoyl-CoA hydratase/isomerase family protein from Hyphomonas neptunium | | Descriptor: | D-MALATE, DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase/isomerase family protein, ... | | Authors: | Szlachta, K, Cooper, D.R, Chapman, H.C, Cymborowski, M.T, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-24 | | Release date: | 2014-03-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Putative enoyl-CoA hydratase/isomerase family protein from Hyphomonas neptunium
To be Published
|
|
4OVT
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM OCHROBACTERIUM ANTHROPI (Oant_3902), TARGET EFI-510153, WITH BOUND L-FUCONATE | | Descriptor: | 6-deoxy-L-galactonic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-14 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O92
 
 | | Crystal structure of a Glutathione S-transferase from Pichia kudriavzevii (Issatchenkia orientalis), target EFI-501747 | | Descriptor: | Glutathione S-transferase, SULFATE ION | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Attonito, J.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-31 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of a Glutathione S-transferase from Pichia kudriavzevii (Issatchenkia orientalis), target EFI-501747
TO BE PUBLISHED
|
|
4OMR
 
 | | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase from Thermobifida fusca YX in complex with acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, BENZAMIDINE, Enoyl-CoA hydratase | | Authors: | Langner, K.M, Cooper, D.R, Chapman, H.C, Handing, K.B, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-27 | | Release date: | 2014-04-02 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase from Thermobifida fusca YX in complex with acetoacetyl-CoA
To be Published
|
|
4OAN
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from rhodopseudomonas palustris HaA2 (RPB_2686), TARGET EFI-510221, with density modeled as (S)-2-hydroxy-2-methyl-3-oxobutanoate ((S)-2-Acetolactate) | | Descriptor: | (2S)-2-hydroxy-2-methyl-3-oxobutanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-01-05 | | Release date: | 2014-01-22 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O9K
 
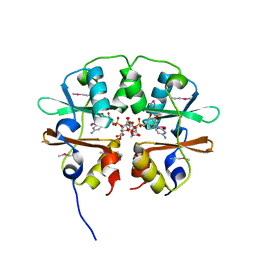 | | Crystal structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Methylococcus capsulatus in complex with CMP-Kdo | | Descriptor: | Arabinose 5-phosphate isomerase, CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL | | Authors: | Shabalin, I.G, Cooper, D.R, Shumilin, I.A, Zimmerman, M.D, Majorek, K.A, Hammonds, J, Hillerich, B.S, Nawar, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and kinetic properties of D-arabinose 5-phosphate isomerase from Methylococcus capsulatus
To be Published
|
|
4OXM
 
 | |
1K4Z
 
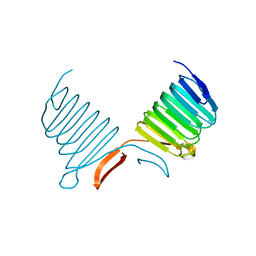 | | C-terminal Domain of Cyclase Associated Protein | | Descriptor: | Adenylyl Cyclase-Associated Protein | | Authors: | Rozwarski, D.A, Fedorov, A.A, Dodatko, T, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-10-09 | | Release date: | 2002-03-13 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the actin binding domain of the cyclase-associated protein
Biochemistry, 43, 2004
|
|
1K8F
 
 | | CRYSTAL STRUCTURE OF THE HUMAN C-TERMINAL CAP1-ADENYLYL CYCLASE ASSOCIATED PROTEIN | | Descriptor: | ADENYLYL CYCLASE-ASSOCIATED PROTEIN | | Authors: | Patskovsky, Y.V, Chance, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-10-24 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the actin binding domain of the cyclase-associated protein.
Biochemistry, 43, 2004
|
|
1LCU
 
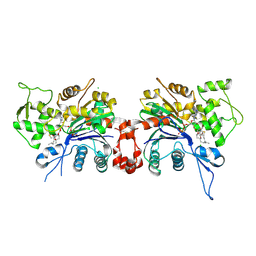 | | Polylysine Induces an Antiparallel Actin Dimer that Nucleates Filament Assembly: Crystal Structure at 3.5 A Resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Bubb, M.R, Govindasamy, L, Yarmola, E.G, Vorobiev, S.M, Almo, S.C, Somasundaram, T, Chapman, M.S, Agbandje-Mckenna, M, Mckenna, R. | | Deposit date: | 2002-04-06 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Polylysine induces an antiparallel actin dimer that nucleates filament assembly: crystal structure at 3.5-A resolution.
J.Biol.Chem., 277, 2002
|
|
1L9G
 
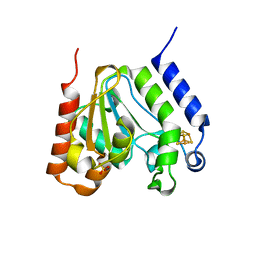 | | CRYSTAL STRUCTURE OF URACIL-DNA GLYCOSYLASE FROM T. MARITIMA | | Descriptor: | Conserved hypothetical protein, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Rajashankar, K.R, Dodatko, T, Thirumuruhan, R.A, Sandigursky, M, Bresnik, A, Chance, M.R, Franklin, W.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-03-22 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of uracil-DNA glycosylase from T. Maritima
To be Published
|
|
1KQ5
 
 | |
1LH0
 
 | | Crystal Structure of Salmonella typhimurium OMP Synthase in Complex with MGPRPP and Orotate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, MAGNESIUM ION, OMP synthase, ... | | Authors: | Fedorov, A.A, Panneerselvam, K, Shi, W, Grubmeyer, C, Almo, S.C. | | Deposit date: | 2002-04-16 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Salmonella typhimurium OMP Synthase in a Complete Substrate Complex.
Biochemistry, 51, 2012
|
|
1L1Q
 
 | | Crystal Structure of APRTase from Giardia lamblia Complexed with 9-deazaadenine | | Descriptor: | 9-DEAZAADENINE, Adenine phosphoribosyltransferase, SULFATE ION | | Authors: | Shi, W, Sarver, A.E, Wang, C.C, Tanaka, K.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-02-19 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Closed Site Complexes of Adenine Phosphoribosyltransferase from Giardia lamblia
Reveal a Mechanism of Ribosyl Migration.
J.Biol.Chem., 277, 2002
|
|
1L1R
 
 | | Crystal Structure of APRTase from Giardia lamblia Complexed with 9-deazaadenine, Mg2+ and PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 9-DEAZAADENINE, Adenine phosphoribosyltransferase, ... | | Authors: | Shi, W, Sarver, A.E, Wang, C.C, Tanaka, K.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-02-19 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Closed Site Complexes of Adenine Phosphoribosyltransferase from Giardia lamblia
Reveal a Mechanism of Ribosyl Migration.
J.Biol.Chem., 277, 2002
|
|
1JUF
 
 | | Structure of Minor Histocompatibility Antigen peptide, H13b, complexed to H2-Db | | Descriptor: | Beta-2-microglobulin, H13b peptide, H2-Db major histocompatibility antigen | | Authors: | Ostrov, D.A, Roden, M.M, Shi, W, Palmieri, E, Christianson, G.J, Mendoza, L, Villaflor, G, Tilley, D, Shastri, N, Grey, H, Almo, S.C, Roopenian, D.C, Nathenson, S.G. | | Deposit date: | 2001-08-24 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How H13 histocompatibility peptides differing by a single methyl group and lacking conventional MHC binding anchor motifs determine self-nonself discrimination.
J.Immunol., 168, 2002
|
|
1M9P
 
 | | Crystalline Human Carbonmonoxy Hemoglobin C Exhibits The R2 Quaternary State at Neutral pH In The Presence of Polyethylene Glycol: The 2.1 Angstrom Resolution Crystal Structure | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Patskovska, L.N, Patskovsky, Y.V, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2002-07-29 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | COHbC and COHbS crystallize in the R2 quaternary state at neutral pH in the presence of PEG 4000.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1K27
 
 | | Crystal Structure of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase in Complex with a Transition State Analogue | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, 5'-Deoxy-5'-Methylthioadenosine Phosphorylase, PHOSPHATE ION | | Authors: | Shi, W, Singh, V, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2001-09-26 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Picomolar transition state analogue inhibitors of human 5'-methylthioadenosine phosphorylase and X-ray structure with MT-immucillin-A
Biochemistry, 43, 2004
|
|
1JHW
 
 | |
1K1K
 
 | | Structure of Mutant Human Carbonmonoxyhemoglobin C (beta E6K) at 2.0 Angstrom Resolution in Phosphate Buffer. | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN ALPHA CHAIN, HEMOGLOBIN BETA CHAIN, ... | | Authors: | Dewan, J.C, Taylor-Feeling, A, Puius, Y.A, Patskovska, L, Patskovsky, Y, Nagel, R.L, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2001-09-25 | | Release date: | 2002-12-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of mutant human carbonmonoxyhemoglobin C (betaE6K) at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
