7TPJ
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, Putative cell surface polysaccharide polymerase/ligase | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
7TPG
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, GERANYL DIPHOSPHATE, ... | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
6U8C
 
 | | Crystal structure of an engineered ultra-high affinity Fab-Protein G complex | | Descriptor: | Antibody heavy chain Fab, Antibody light chain Fab, Protein G | | Authors: | Slezak, T, Filippova, E.V, Davydova, E.K, Kossiakoff, A.A. | | Deposit date: | 2019-09-04 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An engineered ultra-high affinity Fab-Protein G pair enables a modular antibody platform with multifunctional capability.
Protein Sci., 29, 2020
|
|
6UKJ
 
 | | Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Chloroquine resistance transporter, Fab Heavy Chain, ... | | Authors: | Kim, J, Tan, Y.Z, Wicht, K.J, Erramilli, S.K, Dhingra, S.K, Okombo, J, Vendome, J, Hagenah, L.M, Giacometti, S.I, Warren, A.L, Nosol, K, Roepe, P.D, Potter, C.S, Carragher, B, Kossiakoff, A.A, Quick, M, Fidock, D.A, Mancia, F. | | Deposit date: | 2019-10-05 | | Release date: | 2019-12-04 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and drug resistance of the Plasmodium falciparum transporter PfCRT.
Nature, 576, 2019
|
|
6USF
 
 | | CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor with varenicline in complex with anti-BRIL synthetic antibody BAK5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, ... | | Authors: | Alvarez, F.J.D, Mukherjee, S, Han, S, Ammirati, M, Kossiakoff, A.A. | | Deposit date: | 2019-10-26 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
6UR8
 
 | | CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor in complex with varenicline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chimera of soluble cytochrome b562 (BRIL) and neuronal acetylcholine receptor subunit alpha-4, Neuronal acetylcholine receptor subunit beta-2, ... | | Authors: | Alvarez, F.J.D, Mukherjee, S, Han, S, Ammirati, M, Kossiakoff, A.A. | | Deposit date: | 2019-10-22 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
4XH2
 
 | | Crystal structure of human paxillin LD4 motif in complex with Fab fragment | | Descriptor: | ACETATE ION, ACETYL GROUP, Fab Heavy Chain, ... | | Authors: | Nocula-Lugowska, M, Lugowski, M, Salgia, R, Kossiakoff, A.A. | | Deposit date: | 2015-01-04 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering Synthetic Antibody Inhibitors Specific for LD2 or LD4 Motifs of Paxillin.
J.Mol.Biol., 427, 2015
|
|
7PTI
 
 | |
4XWO
 
 | | Structure of Get3 bound to the transmembrane domain of Sec22 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-29 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4XMN
 
 | | Structure of the yeast coat nucleoporin complex, space group P212121 | | Descriptor: | Antibody 87 heavy chain, Antibody 87 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
4XMM
 
 | | Structure of the yeast coat nucleoporin complex, space group C2 | | Descriptor: | Antibody 57 heavy chain, Antibody 57 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (7.384 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
4XGZ
 
 | | Crystal structure of human paxillin LD2 motif in complex with Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, FAB HEAVY CHAIN, FAB LIGHT CHAIN, ... | | Authors: | Nocula-Lugowska, M, Lugowski, M, Salgia, R, Kossiakoff, A.A. | | Deposit date: | 2015-01-04 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineering Synthetic Antibody Inhibitors Specific for LD2 or LD4 Motifs of Paxillin.
J.Mol.Biol., 427, 2015
|
|
7RTH
 
 | | Crystal structure of an anti-lysozyme nanobody in complex with an anti-nanobody Fab "NabFab" | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fragment Antigen-Binding Heavy Chain, ... | | Authors: | Filippova, E.V, Mukherjee, S, Bloch, J.S, Locher, K.P, Kossiakoff, A.A. | | Deposit date: | 2021-08-13 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4XTR
 
 | | Structure of Get3 bound to the transmembrane domain of Pep12 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4XVU
 
 | | Structure of Get3 bound to the transmembrane domain of Nyv1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, Antibody heavy chain, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-28 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
2FCS
 
 | | X-ray Crystal Structure of a Chemically Synthesized [L-Gln35]Ubiquitin with a Cubic Space Group | | Descriptor: | ACETATE ION, CADMIUM ION, SULFATE ION, ... | | Authors: | Bang, D, Gribenko, A.V, Tereshko, V, Kossiakoff, A.A, Kent, S.B, Makhatadze, G.I. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis.
Nat.Chem.Biol., 2, 2006
|
|
2FCN
 
 | | X-ray Crystal Structure of a Chemically Synthesized [D-Val35]Ubiquitin with a Cubic Space Group | | Descriptor: | ACETATE ION, CADMIUM ION, Ubiquitin | | Authors: | Bang, D, Gribenko, A.V, Tereshko, V, Kossiakoff, A.A, Kent, S.B, Makhatadze, G.I. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis.
Nat.Chem.Biol., 2, 2006
|
|
2FCQ
 
 | | X-ray Crystal Structure of a Chemically Synthesized Ubiquitin with a Cubic Space Group | | Descriptor: | CADMIUM ION, Ubiquitin | | Authors: | Bang, D, Gribenko, A.V, Tereshko, V, Kossiakoff, A.A, Kent, S.B, Makhatadze, G.I. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis.
Nat.Chem.Biol., 2, 2006
|
|
2FD7
 
 | | X-ray Crystal Structure of Chemically Synthesized Crambin | | Descriptor: | Crambin | | Authors: | Bang, D, Tereshko, V, Kossiakoff, A.A, Kent, S.B. | | Deposit date: | 2005-12-13 | | Release date: | 2007-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Role of a salt bridge in the model protein crambin explored by chemical protein synthesis: X-ray structure of a unique protein analogue, [V15A]crambin-alpha-carboxamide.
Mol Biosyst, 5, 2009
|
|
2FCM
 
 | | X-ray Crystal Structure of a Chemically Synthesized [D-Gln35]Ubiquitin with a Cubic Space Group | | Descriptor: | ACETATE ION, CADMIUM ION, Ubiquitin | | Authors: | Bang, D, Gribenko, A.V, Tereshko, V, Kossiakoff, A.A, Kent, S.B, Makhatadze, G.I. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis.
Nat.Chem.Biol., 2, 2006
|
|
2FD9
 
 | | X-ray Crystal Structure of Chemically Synthesized Crambin-{alpha}carboxamide | | Descriptor: | Crambin | | Authors: | Bang, D, Tereshko, V, Kossiakoff, A.A, Kent, S.B. | | Deposit date: | 2005-12-13 | | Release date: | 2007-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Role of a salt bridge in the model protein crambin explored by chemical protein synthesis: X-ray structure of a unique protein analogue, [V15A]crambin-alpha-carboxamide.
Mol Biosyst, 5, 2009
|
|
2FXU
 
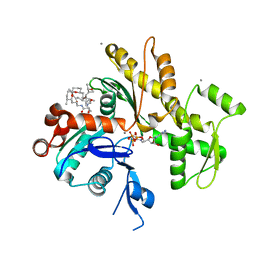 | | X-ray Structure of Bistramide A- Actin Complex at 1.35 A resolution. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rizvi, S.A, Tereshko, V, Kossiakoff, A.A, Kozmin, S.A. | | Deposit date: | 2006-02-06 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of bistramide a-actin complex at a 1.35 A resolution
J.Am.Chem.Soc., 128, 2006
|
|
3HHR
 
 | |
5BJZ
 
 | |
5C8J
 
 | | A YidC-like protein in the archaeal plasma membrane | | Descriptor: | Antibody fragment, heavy chain, light chain, ... | | Authors: | Borowska, M.T, Dominik, P.K, Anghel, S.A, Kossiakoff, A.A, Keenan, R.J. | | Deposit date: | 2015-06-25 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | A YidC-like Protein in the Archaeal Plasma Membrane.
Structure, 23, 2015
|
|
