7TFN
 
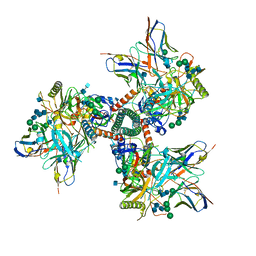 | | Cryo-EM structure of CD4bs antibody Ab1303 in complex with HIV-1 Env trimer BG505 SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-HIV-1 CD4bs antibody Fab Ab1303 - Heavy chain, ... | | Authors: | Yang, Z, Bjorkman, P.J. | | Deposit date: | 2022-01-06 | | Release date: | 2022-01-19 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Neutralizing antibodies induced in immunized macaques recognize the CD4-binding site on an occluded-open HIV-1 envelope trimer.
Nat Commun, 13, 2022
|
|
7TFO
 
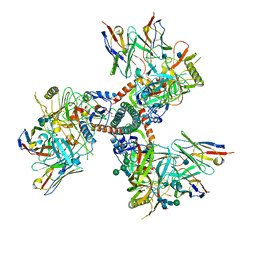 | | Cryo-EM structure of HIV-1 Env trimer BG505 SOSIP.664 in complex with CD4bs antibody Ab1573 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD4 binding site antibody Ab1573 - Fab heavy chain, ... | | Authors: | Yang, Z, Bjorkman, P.J. | | Deposit date: | 2022-01-06 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Neutralizing antibodies induced in immunized macaques recognize the CD4-binding site on an occluded-open HIV-1 envelope trimer.
Nat Commun, 13, 2022
|
|
6CH7
 
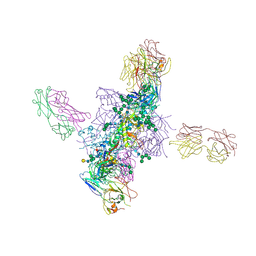 | |
6CH8
 
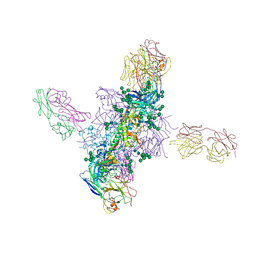 | |
6CH9
 
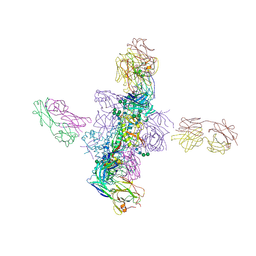 | |
6CHB
 
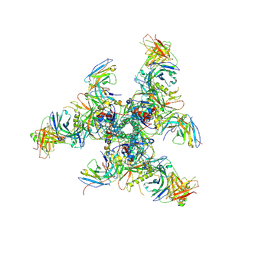 | |
6U0L
 
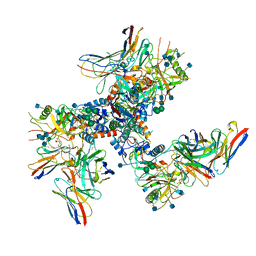 | |
6U0N
 
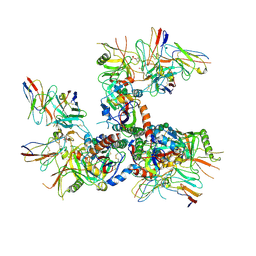 | |
7UGP
 
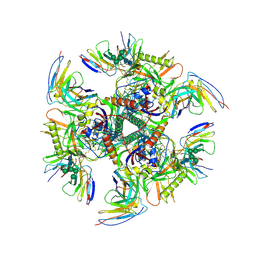 | |
7UGO
 
 | |
7UGQ
 
 | |
7UGN
 
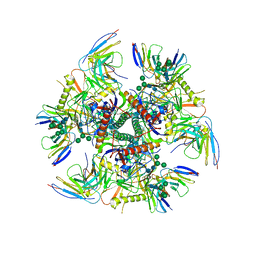 | |
7UGM
 
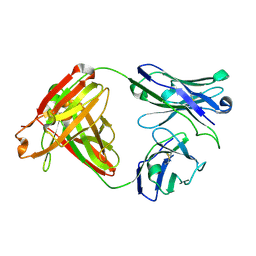 | |
5THR
 
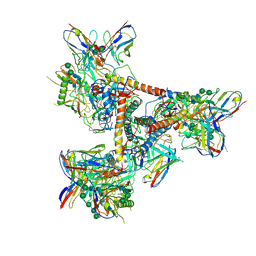 | | Cryo-EM structure of a BG505 Env-sCD4-17b-8ANC195 complex | | Descriptor: | 17b Fab VH domain, 17b Fab VL domain, 8ANC195 G52K5 VH domain, ... | | Authors: | Wang, H, Bjorkman, P.J. | | Deposit date: | 2016-09-30 | | Release date: | 2016-11-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5UD9
 
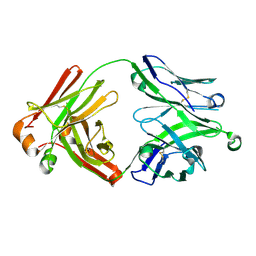 | | Crystal structure of 354BG18 Fab | | Descriptor: | Fab heavy chain, Light chain | | Authors: | Scharf, L, Bjorkman, P.J. | | Deposit date: | 2016-12-23 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller.
Sci Transl Med, 9, 2017
|
|
5VIY
 
 | |
5VJ6
 
 | |
7K8N
 
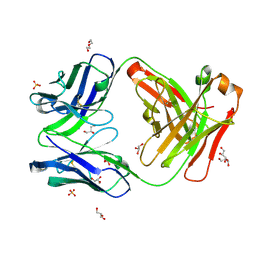 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C102 | | Descriptor: | C102 Fab Heavy Chain, C102 Fab Light Chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8X
 
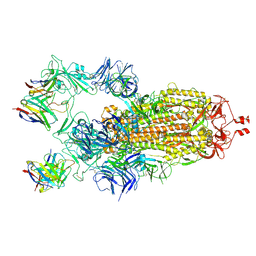 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C121 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C121 Fab Heavy chain, C121 Fab Light chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8Q
 
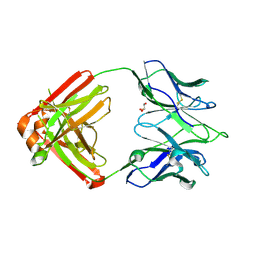 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C121 | | Descriptor: | C121 Fab Heavy Chain, C121 Fab Light Chain, GLYCEROL | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8Y
 
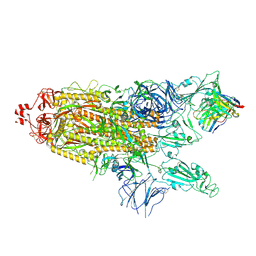 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C121 (State 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C121 Fab Heavy chain, C121 Fab Light chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8P
 
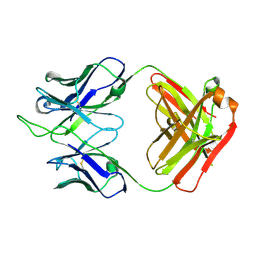 | |
7K8M
 
 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C102 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C102 Fab Heavy Chain, C102 Fab Light Chain, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8V
 
 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C110 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C110 Fab Heavy Chain, ... | | Authors: | Dam, K.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7KDE
 
 | |
