4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4DSC
 
 | | Complex structure of abscisic acid receptor PYL3 with (+)-ABA in spacegroup of H32 at 1.95A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION | | Authors: | Zhang, X, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
2MP0
 
 | | Protein Phosphorylation upon a Fleeting Encounter | | Descriptor: | Glucose-specific phosphotransferase enzyme IIA component, PHOSPHITE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Xing, Q, Yang, J, Huang, P, Zhang, W, Tang, C. | | Deposit date: | 2014-05-08 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Visualizing an ultra-weak protein-protein interaction in phosphorylation signaling.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3AL3
 
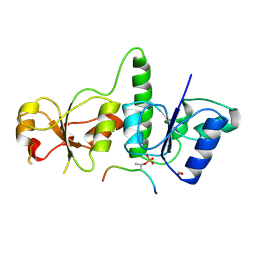 | | Crystal Structure of TopBP1 BRCT7/8-BACH1 peptide complex | | Descriptor: | DNA topoisomerase 2-binding protein 1, FORMIC ACID, Peptide of Fanconi anemia group J protein | | Authors: | Leung, C.C, Glover, J.N. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control
J.Biol.Chem., 286, 2011
|
|
3AL2
 
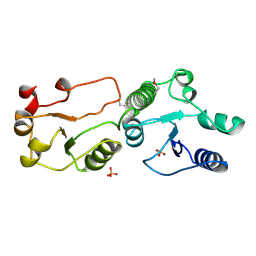 | | Crystal Structure of TopBP1 BRCT7/8 | | Descriptor: | DNA topoisomerase 2-binding protein 1, SULFATE ION | | Authors: | Leung, C.C, Glover, J.N. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control
J.Biol.Chem., 286, 2011
|
|
8Z9Z
 
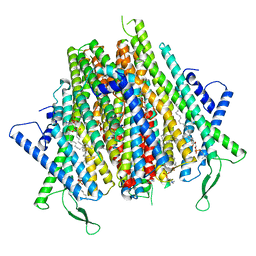 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-24 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
8Z9A
 
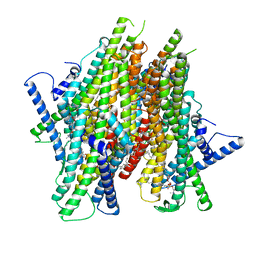 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum bound with geranyl acetate | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
3KL1
 
 | |
3OJI
 
 | | X-ray crystal structure of the Py13 -pyrabactin complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL3, SULFATE ION | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2010-08-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
6KOW
 
 | |
3OQU
 
 | | Crystal structure of native abscisic acid receptor PYL9 with ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL9 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2010-09-04 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural Insights into the Abscisic Acid Stereospecificity by the ABA Receptors PYR/PYL/RCAR
Plos One, 8, 2013
|
|
4DS8
 
 | | Complex structure of abscisic acid receptor PYL3-(+)-ABA-HAB1 in the presence of Mn2+ | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, GLYCEROL, ... | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
5IL1
 
 | | Crystal structure of SAM-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL2
 
 | | Crystal structure of SAH-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL0
 
 | | Crystal structural of the METTL3-METTL14 complex for N6-adenosine methylation | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, METTL14, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
7Y01
 
 | |
5DLK
 
 | | The crystal structure of CT mutant | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-09-06 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
5DIJ
 
 | | The crystal structure of CT | | Descriptor: | CHLORIDE ION, GLYCEROL, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-09-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
5EGF
 
 | | The crystal structure of SeMet-CT | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-10-27 | | Release date: | 2016-10-19 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
5EJD
 
 | | The crystal structure of holo T3CT | | Descriptor: | 4'-PHOSPHOPANTETHEINE, GLYCEROL, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
1GP5
 
 | | Anthocyanidin synthase from Arabidopsis thaliana complexed with trans-dihydroquercetin | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, (2S,3S)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Wilmouth, R.C, Turnbull, J.J, Welford, R.W.D, Clifton, I.J, Prescott, A.G, Schofield, C.J. | | Deposit date: | 2001-10-30 | | Release date: | 2002-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis Thaliana.
Structure, 10, 2002
|
|
4N3Z
 
 | |
4N3X
 
 | |
4N3Y
 
 | |
4Q9U
 
 | | Crystal structure of the Rab5, Rabex-5delta and Rabaptin-5C21 complex | | Descriptor: | Rab GTPase-binding effector protein 1, Rab5 GDP/GTP exchange factor, Ras-related protein Rab-5A | | Authors: | Zhang, Z, Zhang, T, Ding, J. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.618 Å) | | Cite: | Molecular mechanism for Rabex-5 GEF activation by Rabaptin-5
Elife, 3, 2014
|
|
