5SCF
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 99 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-(3-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)glycine, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCB
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 28 | | Descriptor: | (3R)-1-(3,4-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)piperidine-3-carboxylic acid, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCG
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 101 | | Descriptor: | (3R)-1-(3-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)piperidine-3-carboxylic acid, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCL
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 1 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ALIZARIN RED, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCD
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 58 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonic acid, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SC9
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 29 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 1-(3,4-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)piperidine-4-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.685 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCE
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 55 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-amino-4-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonic acid, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCA
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 36 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-[(3R)-1-(3,4-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)piperidine-3-carbonyl]-L-aspartic acid, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCH
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 100 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SC8
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 17 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-[(3,4-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)amino]pentanoic acid, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
6VEO
 
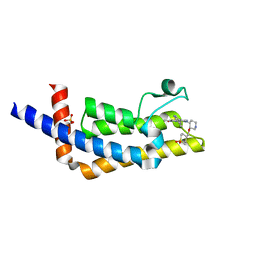 | | ATAD2B bromodomain in complex with 4-({[(3R,4R)-4-{[3-methyl-5-(5-methylpyridin-3-yl)-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl]amino}piperidin-3-yl]oxy}methyl)-1lambda~6~-thiane-1,1-dione (compound 38) | | Descriptor: | 4-({[(3R,4R)-4-{[3-methyl-5-(5-methylpyridin-3-yl)-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl]amino}piperidin-3-yl]oxy}methyl)-1lambda~6~-thiane-1,1-dione, ATPase family AAA domain-containing protein 2B, SULFATE ION | | Authors: | Glass, K.C. | | Deposit date: | 2020-01-02 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Recognition of Mono- and Diacetylated Histones by the ATAD2B Bromodomain.
J.Med.Chem., 63, 2020
|
|
8YQJ
 
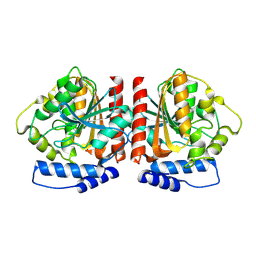 | | Crystal structure of HylD1 | | Descriptor: | Lipase | | Authors: | Wang, N, Li, C.Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular insights into the catalytic mechanism of a phthalate ester hydrolase.
J Hazard Mater, 476, 2024
|
|
1RUR
 
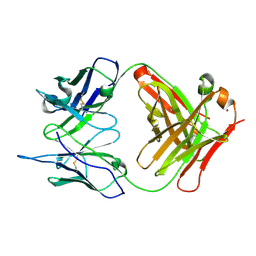 | | Crystal Structure (I) of native Diels-Alder antibody 13G5 Fab at pH 8.0 with a data set collected at SSRL beamline 9-1. | | Descriptor: | ZINC ION, immunoglobulin 13G5, heavy chain, ... | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
1RUQ
 
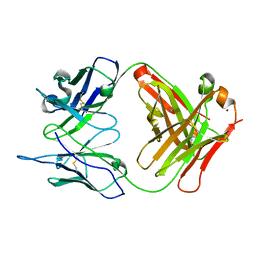 | | Crystal Structure (H) of u.v.-irradiated Diels-Alder antibody 13G5 Fab at pH 8.0 with a data set collected in house. | | Descriptor: | ZINC ION, immunoglobulin 13G5 heavy chain, immunoglobulin 13G5 light chain | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
8IYQ
 
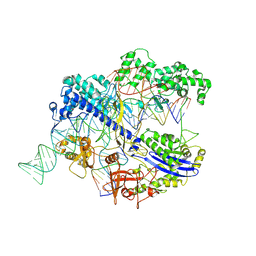 | | Structure of CbCas9 bound to 20-nucleotide complementary DNA substrate | | Descriptor: | NTS, TS, deadCbCas9, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-04-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8HP3
 
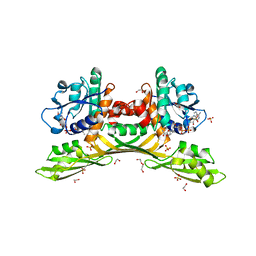 | | Crystal structure of meso-diaminopimelate dehydrogenase from Prevotella timonensis | | Descriptor: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tan, Y, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Rational Design of Meso -Diaminopimelate Dehydrogenase with Enhanced Reductive Amination Activity for Efficient Production of d- p -Hydroxyphenylglycine.
Appl.Environ.Microbiol., 89, 2023
|
|
8HP0
 
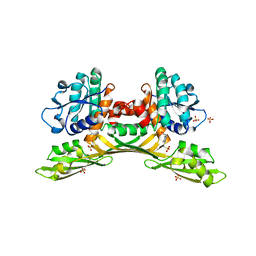 | | Crystal structure of meso-diaminopimelate dehydrogenase from Prevotella timonensis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Meso-diaminopimelate D-dehydrogenase, SULFATE ION | | Authors: | Tan, Y, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Rational Design of Meso -Diaminopimelate Dehydrogenase with Enhanced Reductive Amination Activity for Efficient Production of d- p -Hydroxyphenylglycine.
Appl.Environ.Microbiol., 89, 2023
|
|
8HPE
 
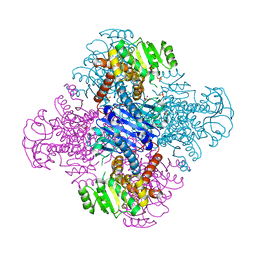 | | Crystal structure of Leucine dehydrogenase | | Descriptor: | GLYCEROL, Leucine dehydrogenase, SULFATE ION | | Authors: | Li, X, Song, W. | | Deposit date: | 2022-12-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A Tri-Enzyme Cascade for Efficient Production of L-2-Aminobutyrate from L-Threonine.
Chembiochem, 24, 2023
|
|
8HR6
 
 | |
4FL6
 
 | | Crystal structure of the complex of the 3-MBT repeat domain of L3MBTL3 and UNC1215 | | Descriptor: | Lethal(3)malignant brain tumor-like protein 3, UNKNOWN ATOM OR ION, [2-(phenylamino)benzene-1,4-diyl]bis{[4-(pyrrolidin-1-yl)piperidin-1-yl]methanone} | | Authors: | Zhong, N, Tempel, W, Ravichandran, M, Dong, A, Ingerman, L.A, Graslund, S, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain.
Nat. Chem. Biol., 9, 2013
|
|
5WZE
 
 | | The structure of Pseudomonas aeruginosa aminopeptidase PepP | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Aminopeptidase P, ... | | Authors: | Bao, R, Peng, C.T, Liu, L, He, L.H, Li, C.C, Li, T, Shen, Y.L, Zhu, Y.B, Song, Y.J. | | Deposit date: | 2017-01-17 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structure-Function Relationship of Aminopeptidase P from Pseudomonas aeruginosa.
Front Microbiol, 8, 2017
|
|
4HSG
 
 | | Crystal structure of human PRMT3 in complex with an allosteric inhibitor (PRMT3- KTD) | | Descriptor: | 1-(1,2,3-benzothiadiazol-6-yl)-3-(2-oxo-2-phenylethyl)urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dobrovetsky, E, Dong, A, Liu, F, Li, F, Tempel, W, Siarheyeva, A, Hajian, T, Smil, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-30 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting an allosteric binding site of PRMT3 yields potent and selective inhibitors.
J. Med. Chem., 56, 2013
|
|
7VVV
 
 | | Crystal structure of MmtN | | Descriptor: | PHOSPHATE ION, SAM-dependent methyltransferase | | Authors: | Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
7VVX
 
 | | MmtN-SAH-Met complex | | Descriptor: | METHIONINE, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhang, Y.Z, Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
7VVW
 
 | | MmtN-SAM complex | | Descriptor: | GLYCEROL, PHOSPHATE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Zhang, Y.Z, Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
