7Z3Q
 
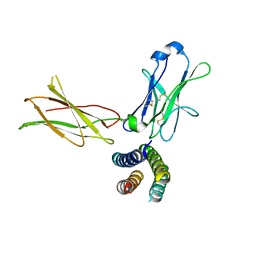 | | Crystal structure of the human leptin:LepR-CRH2 encounter complex to 3.6 A resolution. | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Verschueren, K, Alexandra, T, Savvides, S.N. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.617 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1TEM
 
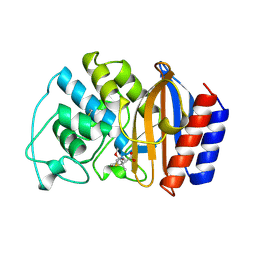 | | 6 ALPHA HYDROXYMETHYL PENICILLOIC ACID ACYLATED ON THE TEM-1 BETA-LACTAMASE FROM ESCHERICHIA COLI | | Descriptor: | 2-(1-CARBOXY-2-HYDROXY-ETHYL)-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, TEM-1 BETA LACTAMASE | | Authors: | Maveyraud, L, Massova, I, Samama, J.P, Mobashery, S. | | Deposit date: | 1996-05-28 | | Release date: | 1997-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of 6Alpha-Hydroxymethylpenicillanate Complexed to the Tem-1 Beta-Lactamase from Escherichia Coli: Evidence on the Mechanism of Action of a Novel Inhibitor Designed by a Computer-Aided Process
J.Am.Chem.Soc., 118, 1996
|
|
4Z9V
 
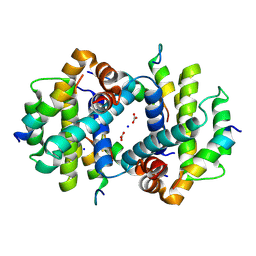 | | TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL | | Descriptor: | BICARBONATE ION, Bcl-2-like protein 1,APOPTOSIS REGULATOR BCL-XL, SODIUM ION, ... | | Authors: | Cura, V, Agez, M, Thebault, S, Cavarelli, J. | | Deposit date: | 2015-04-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL.
Sci Rep, 6, 2016
|
|
1I5N
 
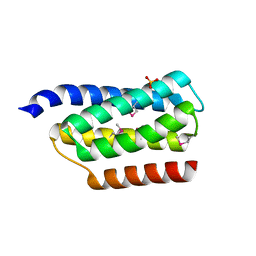 | | Crystal structure of the P1 domain of CheA from Salmonella typhimurium | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, SULFATE ION | | Authors: | Mourey, L, Da Re, S, Pedelacq, J.-D, Tolstyk, T, Faurie, C, Guillet, V, Stock, J.B, Samama, J.-P. | | Deposit date: | 2001-02-28 | | Release date: | 2001-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the CheA histidine phosphotransfer domain that mediates response regulator phosphorylation in bacterial chemotaxis
J.Biol.Chem., 276, 2001
|
|
6Z0L
 
 | | Het-N2 - De novo designed three-helix heterodimer with Cysteine at the N2 position of the alpha-helix | | Descriptor: | CADMIUM ION, Cys-N2 Strand, Positive Strand, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
6Z0M
 
 | | Het-Ncap - De novo designed three-helix heterodimer with Cysteine at the Ncap position of the alpha-helix | | Descriptor: | Cys-Ncap strand, Positive Strand, SULFATE ION, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
7BEY
 
 | | Het-N2-SO3- - De novo designed three-helix heterodimer with Cysteine S-sulfate at the N2 position of the alpha-helix | | Descriptor: | 'Cys-N2-SO3-' Strand, 'Positive' Strand, SULFATE ION | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
1AVB
 
 | | ARCELIN-1 FROM PHASEOLUS VULGARIS L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARCELIN-1, ... | | Authors: | Mourey, L, Pedelacq, J.D, Fabre, C, Rouge, P, Samama, J.P. | | Deposit date: | 1997-09-15 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the arcelin-1 dimer from Phaseolus vulgaris at 1.9-A resolution.
J.Biol.Chem., 273, 1998
|
|
8AV2
 
 | | Crystal structure for the FnIII module of mouse LEP-R in complex with the anti-LEP-R nanobody VHH-4.80 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leptin receptor, SULFATE ION, ... | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVC
 
 | | Mouse leptin:LEP-R complex cryoEM structure (3:3 model) | | Descriptor: | Leptin, Leptin receptor, NICKEL (II) ION | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVD
 
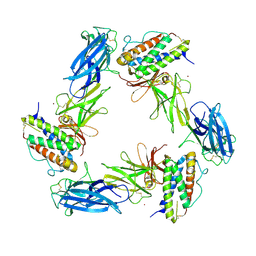 | | Cryo-EM structure for a 3:3 complex between mouse leptin and the mouse LEP-R ectodomain (local refinement) | | Descriptor: | Leptin, Leptin receptor, NICKEL (II) ION | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVO
 
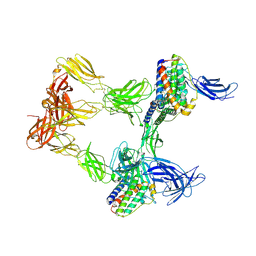 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (open 3:3 model). | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (6.84 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVE
 
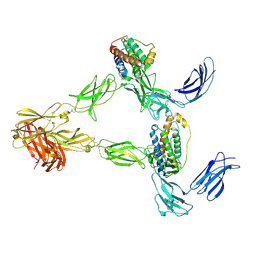 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (2:2 model) | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (5.62 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVF
 
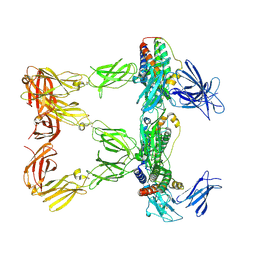 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (closed 3:3 model) | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (6.45 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVB
 
 | | Cryo-EM structure for mouse leptin in complex with the mouse LEP-R ectodomain (1:2 mLEP:mLEPR model). | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8B7Q
 
 | | Cryo-EM structure for the mouse LEPR-CRH2:Leptin:LEPR-Ig complex following symmetry expansion in combination with local refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-09-30 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3HLW
 
 | | CTX-M-9 S70G in complex with cefotaxime | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, CTX-M-9 extended-spectrum beta-lactamase | | Authors: | Delmas, J, Leyssne, D, Dubois, D, Vazeille, E, Robin, F, Bonnet, R. | | Deposit date: | 2009-05-28 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into substrate recognition and product expulsion in CTX-M enzymes.
J.Mol.Biol., 400, 2010
|
|
3HRE
 
 | | X-ray crystallographic structure of CTX-M-9 S70G | | Descriptor: | CTX-M-9 extended-spectrum beta-lactamase, PHOSPHATE ION | | Authors: | Delmas, J, Leyssene, D, Dubois, D, Vazeille, E, Robin, F, Bonnet, R. | | Deposit date: | 2009-06-09 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into substrate recognition and product expulsion in CTX-M enzymes.
J.Mol.Biol., 400, 2010
|
|
3HUO
 
 | | X-ray crystallographic structure of CTX-M-9 S70G in complex with benzylpenicillin | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CTX-M-9 extended-spectrum beta-lactamase, PENICILLIN G | | Authors: | Delmas, J, Leyssene, D, Dubois, D, Robin, F, Bonnet, R. | | Deposit date: | 2009-06-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dynamic view of the early and late steps of the catalytic mechanism mediated by the emerging enzymes CTX-M.
To be Published
|
|
3HVF
 
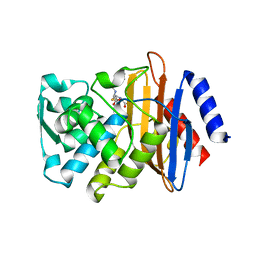 | | X-ray crystallographic structure of CTX-M-9 S70G in complex with hydrolyzed benzylpenicillin | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CTX-M-9 extended-spectrum beta-lactamase | | Authors: | Delmas, J, Leyssene, D, Dubois, D, Vazeille, E, Robin, F, Bonnet, R. | | Deposit date: | 2009-06-16 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into substrate recognition and product expulsion in CTX-M enzymes.
J.Mol.Biol., 400, 2010
|
|
4CN2
 
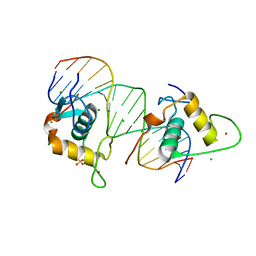 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human Ramp2 Response Element | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*AP*TP*TP*GP*AP*CP*CP*CP*TP*TP*GP*AP*AP*DC *TP*CP*AP)-3', 5'-D(*TP*GP*AP*GP*TP*TP*CP*AP*AP*GP*GP*GP*TP*DC *AP*AP*TP)-3', ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2014-01-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | Structural Basis of Natural Promoter Recognition by the Retinoid X Nuclear Receptor.
Sci.Rep., 5, 2015
|
|
4CN7
 
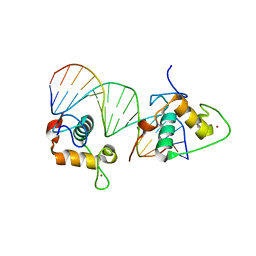 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to an idealized DR1 Response Element | | Descriptor: | 5'-D(*CP*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP *AP*GP)-3', 5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP *AP*GP)-3', CHLORIDE ION, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2014-01-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Basis of Natural Promoter Recognition by the Retinoid X Nuclear Receptor.
Sci.Rep., 5, 2015
|
|
4CN3
 
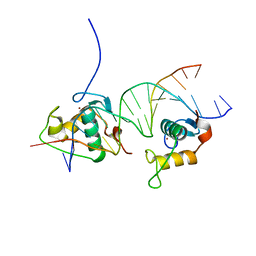 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human Gde1SpA Response Element | | Descriptor: | 5'-D(*CP*TP*AP*GP*TP*TP*CP*AP*AP*AP*GP*TP*TP*CP *AP*CP*A)-3', 5'-D(*TP*GP*TP*GP*AP*AP*CP*TP*TP*TP*GP*AP*AP*CP *TP*AP*G)-3', RETINOIC ACID RECEPTOR RXR-ALPHA, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2014-01-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Natural Promoter Recognition by the Retinoid X Nuclear Receptor.
Sci.Rep., 5, 2015
|
|
4CN5
 
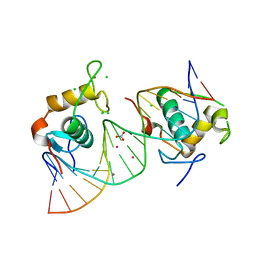 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human Nr1d1 Response Element | | Descriptor: | 5'-D(*AP*TP*TP*GP*AP*AP*CP*TP*CP*TP*GP*AP*CP*CP *CP*CP*AP)-3', 5'-D(*TP*GP*GP*GP*GP*TP*CP*AP*GP*AP*GP*TP*TP*CP *AP*AP*TP)-3', CHLORIDE ION, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2014-01-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Natural Promoter Recognition by the Retinoid X Nuclear Receptor.
Sci.Rep., 5, 2015
|
|
1FQW
 
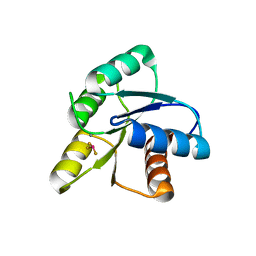 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-09-07 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of activated CheY. Comparison with other activated receiver domains.
J.Biol.Chem., 276, 2001
|
|
