4OCI
 
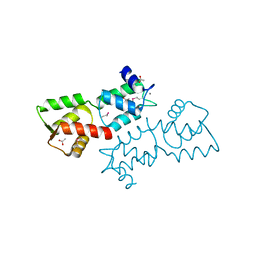 | | Crystal Structure of Calcium Binding Protein-5 from Entamoeba histolytica and its involvement in initiation of phagocytosis of human erythrocytes | | Descriptor: | ACETATE ION, CALCIUM ION, Calmodulin, ... | | Authors: | Kumar, S, Manjasetty, A.B, Zaidi, R, Gourinath, S. | | Deposit date: | 2014-01-09 | | Release date: | 2014-12-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Crystal Structure of Calcium Binding Protein-5 from Entamoeba histolytica and Its Involvement in Initiation of Phagocytosis of Human Erythrocytes.
Plos Pathog., 10, 2014
|
|
5VA4
 
 | |
5NL6
 
 | |
1L7B
 
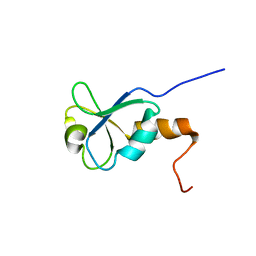 | | Solution NMR Structure of BRCT Domain of T. Thermophilus: Northeast Structural Genomics Consortium Target WR64TT | | Descriptor: | DNA LIGASE | | Authors: | Sahota, G, Dixon, B.L, Huang, Y.P, Aramini, J, Monleon, D, Bhattacharya, D, Swapna, G.V.T, Yin, C, Xiao, R, Anderson, S, Tejero, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-03-14 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Brct Domain from Thermus Thermophilus DNA Ligase
To be Published
|
|
4LY4
 
 | |
3CXC
 
 | | The structure of an enhanced oxazolidinone inhibitor bound to the 50S ribosomal subunit of H. marismortui | | Descriptor: | (3Z)-N-[(4E)-5-(4-{(5S)-5-[(acetylamino)methyl]-2-oxo-1,3-oxazolidin-3-yl}-2-fluorophenyl)pent-4-en-1-yl]-3-(4-methyl-2,6-dioxo-1,6-dihydropyrimidin-5(2H)-ylidene)propanamide, 23S RIBOSOMAL RNA, 5'-R(*CP*CP*A)-3', ... | | Authors: | Ippolito, J.A, Wang, D, Kanyo, Z.F, Duffy, E.M. | | Deposit date: | 2008-04-24 | | Release date: | 2009-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design at the atomic level: design of biaryloxazolidinones as potent orally active antibiotics.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6SL7
 
 | |
6SL3
 
 | |
6SL2
 
 | | ALPHA-ACTININ FROM ENTAMOEBA HISTOLYTICA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Calponin homology domain protein putative, ... | | Authors: | Pinotsis, N, Khan, M.B, Djinovic-Carugo, K. | | Deposit date: | 2019-08-18 | | Release date: | 2020-08-26 | | Last modified: | 2020-11-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Calcium modulates the domain flexibility and function of an alpha-actinin similar to the ancestral alpha-actinin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6U6Z
 
 | | Human SAMHD1 bound to deoxyribo(TG*TTCA)-oligonucleotide | | Descriptor: | DNA polymer TG(PST)TCA, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ZINC ION | | Authors: | Taylor, A.B, Yu, C.H, Ivanov, D.N. | | Deposit date: | 2019-08-30 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification.
Nat Commun, 12, 2021
|
|
6HF2
 
 | | The structure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glycosyl hydrolase family 26 | | Authors: | Bagenholm, V, Logan, D.T, Stalbrand, H. | | Deposit date: | 2018-08-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A surface-exposed GH26 beta-mannanase fromBacteroides ovatus: Structure, role, and phylogenetic analysis ofBoMan26B.
J.Biol.Chem., 294, 2019
|
|
6HF4
 
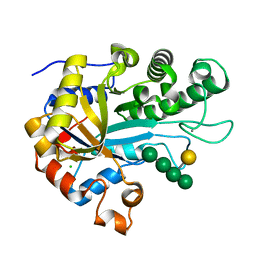 | | The structure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus, complexed with G1M4 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glycosyl hydrolase family 26, ... | | Authors: | Bagenholm, V, Logan, D.T, Stalbrand, H. | | Deposit date: | 2018-08-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | A surface-exposed GH26 beta-mannanase fromBacteroides ovatus: Structure, role, and phylogenetic analysis ofBoMan26B.
J.Biol.Chem., 294, 2019
|
|
7LD3
 
 | | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist and an allosteric ligand | | Descriptor: | ADENOSINE, Chimera protein of Muscarinic acetylcholine receptor M4 and Adenosine receptor A1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Draper-Joyce, C.J, Danev, R, Thal, D.M, Christopoulos, A, Glukhova, A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Positive allosteric mechanisms of adenosine A 1 receptor-mediated analgesia.
Nature, 597, 2021
|
|
7LD4
 
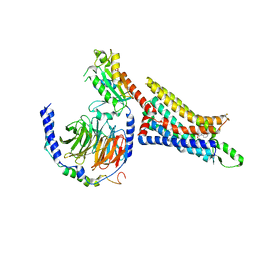 | | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist | | Descriptor: | ADENOSINE, Chimera protein of Muscarinic acetylcholine receptor M4 and Adenosine receptor A1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Draper-Joyce, C.J, Danev, R, Thal, D.M, Christopoulos, A, Glukhova, A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Positive allosteric mechanisms of adenosine A 1 receptor-mediated analgesia.
Nature, 597, 2021
|
|
6C61
 
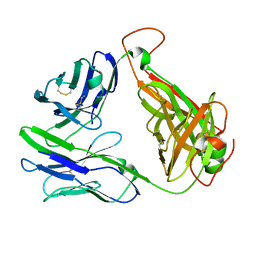 | | MHC-independent T-cell receptor B12A | | Descriptor: | T-cell receptor alpha chain, T-cell receptor beta chain | | Authors: | Lu, J, Sun, P. | | Deposit date: | 2018-01-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of MHC-Independent TCRs and Their Recognition of Native Antigen CD155.
J Immunol., 204, 2020
|
|
6N5W
 
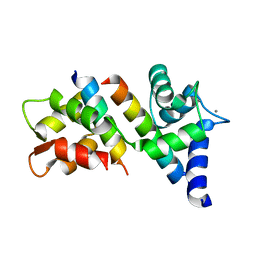 | |
7ZUD
 
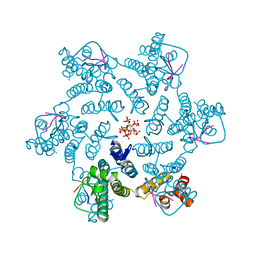 | | Crystal structure of HIV-1 capsid IP6-CPSF6 complex | | Descriptor: | Capsid protein p24, Cleavage and polyadenylation specificity factor subunit 6, INOSITOL HEXAKISPHOSPHATE | | Authors: | Nicastro, G, Taylor, I.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | CP-MAS and Solution NMR Studies of Allosteric Communication in CA-assemblies of HIV-1.
J.Mol.Biol., 434, 2022
|
|
1XN8
 
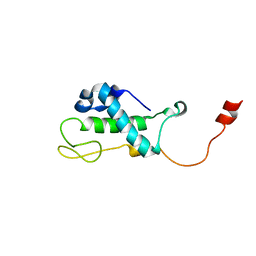 | | Solution Structure of Bacillus subtilis Protein yqbG: The Northeast Structural Genomics Consortium Target SR215 | | Descriptor: | Hypothetical protein yqbG | | Authors: | Liu, G, Ma, L, Shen, Y, Acton, T, Atreya, H.S, Xiao, R, Joachimiak, A, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR data collection and analysis protocol for high-throughput protein structure determination.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5B7X
 
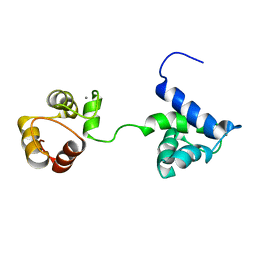 | |
3I3L
 
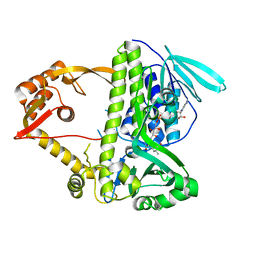 | | Crystal structure of CmlS, a flavin-dependent halogenase | | Descriptor: | Alkylhalidase CmlS, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Podzelinska, K, Soares, A, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-06-30 | | Release date: | 2010-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chloramphenicol Biosynthesis: The Structure of CmlS, a Flavin-Dependent Halogenase Showing a Covalent Flavin-Aspartate Bond
J.Mol.Biol., 397, 2010
|
|
1XNE
 
 | | Solution Structure of Pyrococcus furiosus Protein PF0470: The Northeast Structural Genomics Consortium Target PfR14 | | Descriptor: | hypothetical protein PF0469 | | Authors: | Liu, G, Xiao, R, Parish, D, Ma, L, Sukumaran, D, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR data collection and analysis protocol for high-throughput protein structure determination.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1XN6
 
 | | Solution Structure of Northeast Structural Genomics Target Protein BcR68 encoded in gene Q816V6 of B. cereus | | Descriptor: | hypothetical protein BC4709 | | Authors: | Liu, G, Acton, T, Parish, D, Ma, L, Xu, D, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR data collection and analysis protocol for high-throughput protein structure determination.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5NL7
 
 | | The crystal structure of the Actin Binding Domain (ABD) of alpha actinin from Entamoeba histolytica | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Calponin homology domain protein putative | | Authors: | Pinotsis, N, Djinovic-Carugo, K, Khan, M.B. | | Deposit date: | 2017-04-04 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Calcium modulates the domain flexibility and function of an alpha-actinin similar to the ancestral alpha-actinin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YK8
 
 | | OTU-like deubiquitinase from Legionella -Lpg2529 | | Descriptor: | PLATINUM (II) ION, Uncharacterized protein | | Authors: | Shin, D, Dikic, I. | | Deposit date: | 2020-04-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Bacterial OTU deubiquitinases regulate substrate ubiquitination upon Legionella infection.
Elife, 9, 2020
|
|
6M1X
 
 | |
