9LRE
 
 | | Cryo-EM structure of the histamine H4 receptor-Gi protein complex (Overall) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Matsuzaki, Y, Sano, F.K, Oshima, H.S, Akasaka, H, Kobayashi, K, Tanaka, T, Itoh, Y, Shihoya, W, Kise, Y, Kusakizako, T, Nureki, O. | | Deposit date: | 2025-01-30 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into ligand recognition and G protein preferences across histamine receptor
To Be Published
|
|
9LRB
 
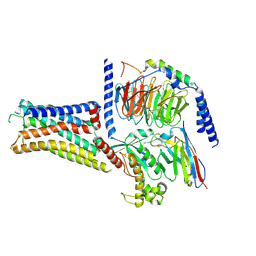 | | Cryo-EM structure of the histamine H1 receptor-Gs protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Matsuzaki, Y, Sano, F.K, Oshima, H.S, Akasaka, H, Kobayashi, K, Tanaka, T, Itoh, Y, Shihoya, W, Kise, Y, Kusakizako, T, Nureki, O. | | Deposit date: | 2025-01-30 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into ligand recognition and G protein preferences across histamine receptors
To Be Published
|
|
9LRD
 
 | | Cryo-EM structure of the histamine H1 receptor-Gi protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Matsuzaki, Y, Sano, F.K, Oshima, H.S, Akasaka, H, Kobayashi, K, Tanaka, T, Itoh, Y, Shihoya, W, Kise, Y, Kusakizako, T, Nureki, O. | | Deposit date: | 2025-01-30 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and G protein preferences across histamine receptors
To Be Published
|
|
9LRC
 
 | | Cryo-EM structure of the histamine H4 receptor-Gi protein complex (Receptor focused) | | Descriptor: | HISTAMINE, Histamine H4 receptor,Genome polyprotein | | Authors: | Matsuzaki, Y, Sano, F.K, Oshima, H.S, Akasaka, H, Kobayashi, K, Tanaka, T, Itoh, Y, Shihoya, W, Kise, Y, Kusakizako, T, Nureki, O. | | Deposit date: | 2025-01-30 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into ligand recognition and G protein preferences across histamine receptors
To Be Published
|
|
5XW6
 
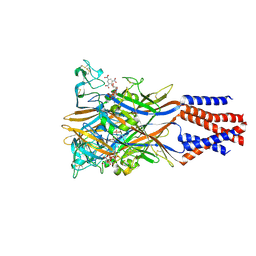 | | Crystal structure of the chicken ATP-gated P2X7 receptor channel in the presence of competitive antagonist TNP-ATP at 3.1 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, P2X purinoceptor, SPIRO(2,4,6-TRINITROBENZENE[1,2A]-2O',3O'-METHYLENE-ADENINE-TRIPHOSPHATE | | Authors: | Kasuya, G, Hattori, M, Nureki, O. | | Deposit date: | 2017-06-29 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into the competitive inhibition of the ATP-gated P2X receptor channel
Nat Commun, 8, 2017
|
|
1QQT
 
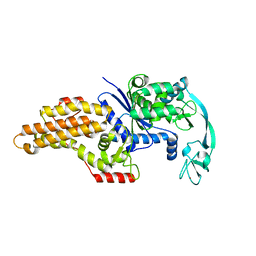 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Mechulam, Y, Schmitt, E, Maveyraud, L, Zelwer, C, Nureki, O, Yokoyama, S, Konno, M, Blanquet, S. | | Deposit date: | 1999-06-08 | | Release date: | 2000-01-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features.
J.Mol.Biol., 294, 1999
|
|
5CZZ
 
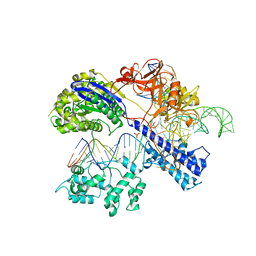 | | Crystal structure of Staphylococcus aureus Cas9 in complex with sgRNA and target DNA (TTGAAT PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Staphylococcus aureus Cas9.
Cell, 162, 2015
|
|
5D4J
 
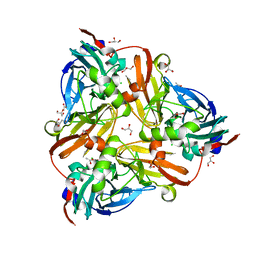 | | Chloride-bound form of a copper nitrite reductase from Alcaligenes faecals | | Descriptor: | ACETIC ACID, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D4H
 
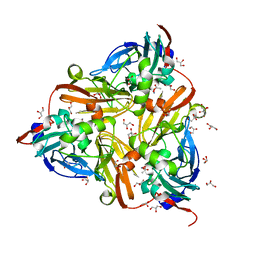 | | High-resolution nitrite complex of a copper nitrite reductase determined by synchrotron radiation crystallography | | Descriptor: | ACETIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2RRN
 
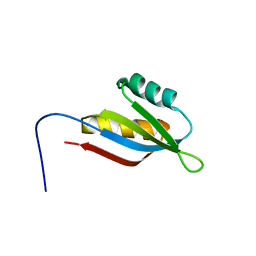 | | Solution structure of SecDF periplasmic domain P4 | | Descriptor: | Probable SecDF protein-export membrane protein | | Authors: | Tanaka, T, Tsukazaki, T, Echizen, Y, Nureki, O, Kohno, T. | | Deposit date: | 2011-01-30 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a membrane component SecDF that enhances protein export
Nature, 474, 2011
|
|
9JTQ
 
 | | Crystal structure of the light-driven proton pump heimdallarchaeial rhodopsin HeimdallR1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, NITRATE ION, RETINAL, ... | | Authors: | Matsuzaki, Y, Tzlil, G, Del Carmen Marin, M, Shihoya, W, Beja, O, Nureki, O. | | Deposit date: | 2024-10-06 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Light-harvesting by antenna-containing rhodopsins in pelagic Asgard archaea
To Be Published
|
|
1A8H
 
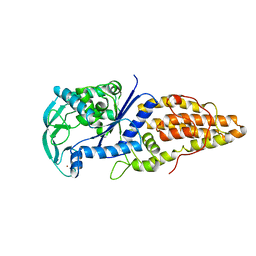 | | METHIONYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS | | Descriptor: | METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sugiura, I, Nureki, O, Ugaji, Y, Kuwabara, S, Lober, B, Giege, R, Moras, D, Yokoyama, S, Konno, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules.
Structure, 8, 2000
|
|
1B7F
 
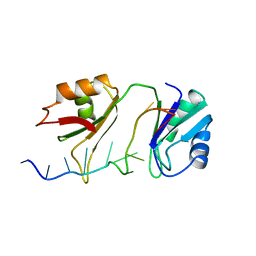 | | SXL-LETHAL PROTEIN/RNA COMPLEX | | Descriptor: | PROTEIN (SXL-LETHAL PROTEIN), RNA (5'-R(P*GP*UP*UP*GP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Handa, N, Nureki, O, Kurimoto, K, Kim, I, Sakamoto, H, Shimura, Y, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-01-23 | | Release date: | 1999-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein.
Nature, 398, 1999
|
|
5XUZ
 
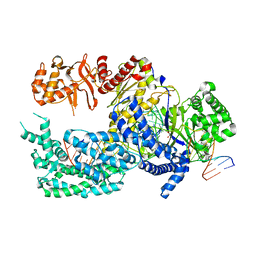 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (CCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*CP*CP*CP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
5XUT
 
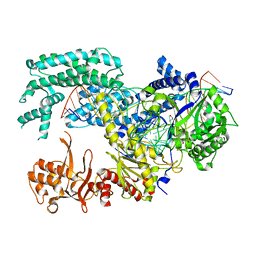 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TCTA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*CP*TP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
5XUS
 
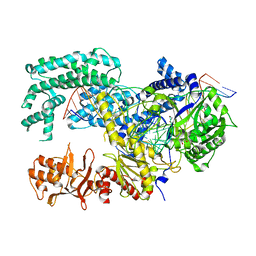 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TTTA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*TP*TP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
5XH6
 
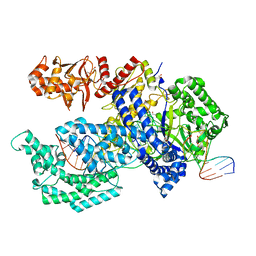 | | Crystal structure of the Acidaminococcus sp. BV3L6 Cpf1 RVR variant in complex with crRNA and target DNA (TATA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CRISPR-associated endonuclease Cpf1, ... | | Authors: | Nishimasu, H, Yamano, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Mol. Cell, 67, 2017
|
|
5XUU
 
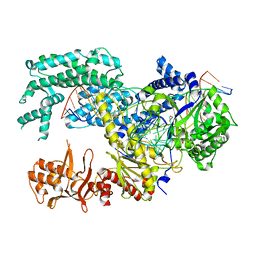 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*CP*CP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
5Y50
 
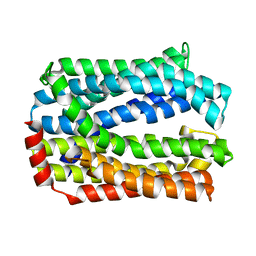 | | Crystal structure of eukaryotic MATE transporter AtDTX14 | | Descriptor: | Protein DETOXIFICATION 14 | | Authors: | Miyauchi, H, Kusakizako, T, Nishizawa, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for xenobiotic extrusion by eukaryotic MATE transporter
Nat Commun, 8, 2017
|
|
2CSX
 
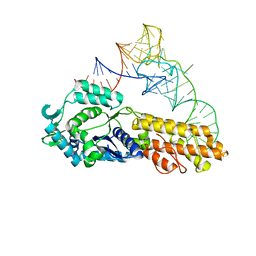 | | Crystal structure of Aquifex aeolicus methionyl-tRNA synthetase complexed with tRNA(Met) | | Descriptor: | Methionyl-tRNA synthetase, RNA (75-MER) | | Authors: | Nakanishi, K, Ogiso, Y, Nakama, T, Fukai, S, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for anticodon recognition by methionyl-tRNA synthetase.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2CT8
 
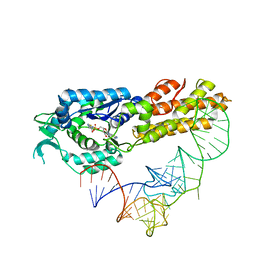 | | Crystal structure of Aquifex aeolicus methionyl-tRNA synthetase complexed with tRNA(Met) and methionyl-adenylate anologue | | Descriptor: | 5'-O-[(L-METHIONYL)-SULPHAMOYL]ADENOSINE, Methionyl-tRNA synthetase, RNA (74-MER) | | Authors: | Nakanishi, K, Ogiso, Y, Nakama, T, Fukai, S, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for anticodon recognition by methionyl-tRNA synthetase.
Nat.Struct.Mol.Biol., 12, 2005
|
|
6LGP
 
 | | cryo-EM structure of TRPV3 in lipid nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, DIUNDECYL PHOSPHATIDYL CHOLINE, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Shimada, H, Kusakizako, T, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-05 | | Release date: | 2020-06-24 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of lipid nanodisc-reconstituted TRPV3 reveals the gating mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LPK
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 293 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
5NDC
 
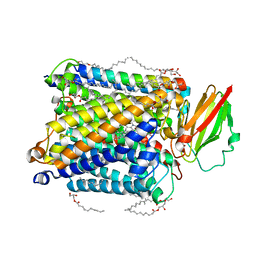 | | Structure of ba3-type cytochrome c oxidase from Thermus thermophilus by serial femtosecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide IIA, ... | | Authors: | Andersson, R, Safari, C, Dods, R, Nango, E, Tanaka, R, Nakane, T, Tono, K, Joti, Y, Bath, P, Dunevall, E, Bosman, E, Nureki, O, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serial femtosecond crystallography structure of cytochrome c oxidase at room temperature.
Sci Rep, 7, 2017
|
|
6LMU
 
 | | Cryo-EM structure of the human CALHM2 | | Descriptor: | Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
