7RGW
 
 | |
7TRM
 
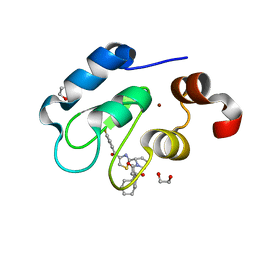 | | Crystal structure of human BIRC2 BIR3 domain in complex with inhibitor LCL-161 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, LCL-161, ... | | Authors: | Tencer, A.H, Klein, B.J, Kutateladze, T.G. | | Deposit date: | 2022-01-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for nuclear accumulation and targeting of the inhibitor of apoptosis BIRC2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TRL
 
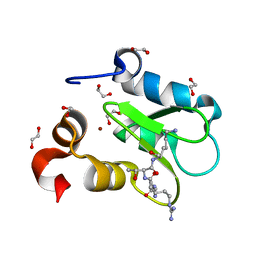 | | Crystal structure of human BIRC2 BIR3 domain in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, Histone H3, ... | | Authors: | Klein, B.J, Tencer, A.H, Kutateladze, T.G. | | Deposit date: | 2022-01-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Molecular basis for nuclear accumulation and targeting of the inhibitor of apoptosis BIRC2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7UHE
 
 | |
4L58
 
 | |
4L7X
 
 | |
5SVX
 
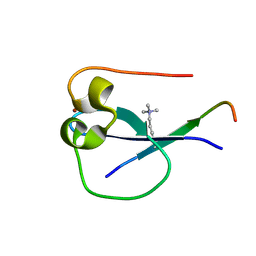 | |
5SVY
 
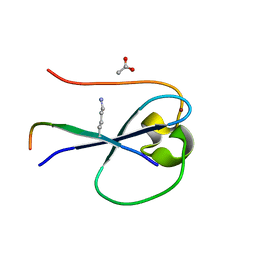 | | MORC3 CW in complex with histone H3K4me1 | | Descriptor: | ACETATE ION, H3K4me1, MORC family CW-type zinc finger protein 3, ... | | Authors: | Tong, Q, Andrews, F.H, Kutateladze, T.G. | | Deposit date: | 2016-08-08 | | Release date: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Multivalent Chromatin Engagement and Inter-domain Crosstalk Regulate MORC3 ATPase.
Cell Rep, 16, 2016
|
|
5SVI
 
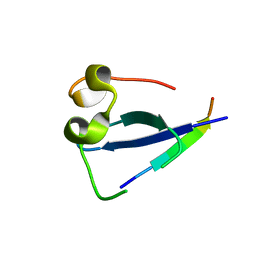 | |
2PNX
 
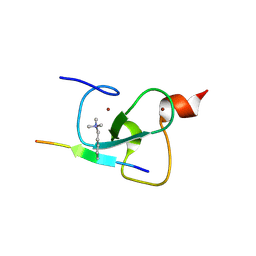 | |
5TAB
 
 | |
5TDW
 
 | | Set3 PHD finger in complex with histone H3K4me3 | | Descriptor: | SET domain-containing protein 3, SODIUM ION, ZINC ION, ... | | Authors: | Andrews, F.H, Ali, M, Kutateladze, T.G. | | Deposit date: | 2016-09-19 | | Release date: | 2016-10-19 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight into Recognition of Methylated Histone H3K4 by Set3.
J. Mol. Biol., 429, 2017
|
|
7TV0
 
 | |
7TUQ
 
 | |
6MIN
 
 | |
6MIM
 
 | |
6MIO
 
 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9pr | | Descriptor: | Histone H3K9pr, Transcription initiation factor TFIID subunit 14 | | Authors: | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6MIQ
 
 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9bu | | Descriptor: | Histone H3K9bu, Transcription initiation factor TFIID subunit 14 | | Authors: | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6MIP
 
 | |
6MIL
 
 | | Crystal structure of AF9 YEATS domain in complex with histone H3K9bu | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histone H3K9bu, MALONATE ION, ... | | Authors: | Vann, K.R, Klein, B.J, Kutateladze, T.G. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6OIE
 
 | |
3LH0
 
 | |
3LGF
 
 | |
3LGL
 
 | |
3MDF
 
 | | Crystal structure of the RRM domain of Cyclophilin 33 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Hom, R.A, Chang, P.Y, Roy, S, Mussleman, C.A, Glass, K.C, Seleznevia, A.I, Gozani, O, Ismagilov, R.F, Cleary, M.L, Kutateladze, T.G. | | Deposit date: | 2010-03-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular mechanism of MLL PHD3 and RNA recognition by the Cyp33 RRM domain.
J.Mol.Biol., 400, 2010
|
|
