7E4T
 
 | | Human TRPC5 apo state structure at 3 angstrom | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
7D4P
 
 | | Structure of human TRPC5 in complex with clemizole | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-[(4-chlorophenyl)methyl]-2-(pyrrolidin-1-ylmethyl)benzimidazole, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
7D4Q
 
 | | Structure of human TRPC5 in complex with HC-070 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 8-(3-chloranylphenoxy)-7-[(4-chlorophenyl)methyl]-3-methyl-1-(3-oxidanylpropyl)purine-2,6-dione, CALCIUM ION, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
8KA5
 
 | |
8KA3
 
 | |
8KA4
 
 | |
8IS5
 
 | |
8IS4
 
 | | Structure of an Isocytosine specific deaminase Vcz in complexed with 5-FU | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-FLUOROURACIL, GLYCEROL, ... | | Authors: | Guo, W.T, Li, X.J, Wu, B.X. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of an isocytosine-specific deaminase VCZ reveals its application potential in the anti-cancer therapy.
Iscience, 26, 2023
|
|
7WW2
 
 | | Structure of an Isocytosine specific deaminase Vcz | | Descriptor: | 8-oxoguanine deaminase, ZINC ION | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-02-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of an isocytosine-specific deaminase VCZ reveals its application potential in the anti-cancer therapy.
Iscience, 26, 2023
|
|
7THS
 
 | | Macrocyclic plasmin inhibitor | | Descriptor: | (2S)-butane-1,2-diol, (6S,9R,20R,23S)-N-{[4-(aminomethyl)phenyl]methyl}-20-[(benzenesulfonyl)amino]-3,13,21-trioxo-2,6,9,14,22-pentaazatetracyclo[23.2.2.2~6,9~.2~15,18~]tritriaconta-1(27),15,17,25,28,30-hexaene-23-carboxamide, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-01-12 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Structural Characterization of Macrocyclic Plasmin Inhibitors.
Chemmedchem, 18, 2023
|
|
7UAH
 
 | | Macrocyclic plasmin inhibitor | | Descriptor: | (2~{R})-butane-1,2-diol, (6S,9R,19S,22R)-N-{[4-(aminomethyl)phenyl]methyl}-22-[(3-chlorobenzene-1-sulfonyl)amino]-3,12,21-trioxo-2,6,9,13,20-pentaazatetracyclo[22.2.2.2~6,9~.2~14,17~]dotriaconta-1(26),14,16,24,27,29-hexaene-19-carboxamide, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-03-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Synthesis and Structural Characterization of Macrocyclic Plasmin Inhibitors.
Chemmedchem, 18, 2023
|
|
5LG4
 
 | |
5M4Y
 
 | |
3GBZ
 
 | |
8F1G
 
 | | Crystal structure of human WDR5 in complex with compound WM662 | | Descriptor: | (2S)-2-({(2S)-3-(3'-chloro[1,1'-biphenyl]-4-yl)-1-oxo-1-[(1H-tetrazol-5-yl)amino]propan-2-yl}oxy)propanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Liu, H. | | Deposit date: | 2022-11-05 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of WDR5-MYC Interaction.
Acs Chem.Biol., 18, 2023
|
|
3GC0
 
 | |
8GMO
 
 | |
4RL5
 
 | |
8Z2O
 
 | |
8Z2N
 
 | |
8Z2M
 
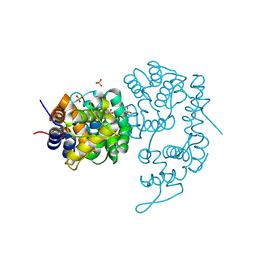 | |
8Z79
 
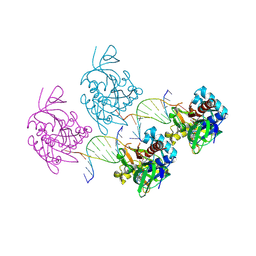 | |
3HIE
 
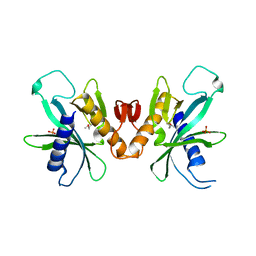 | |
3GTD
 
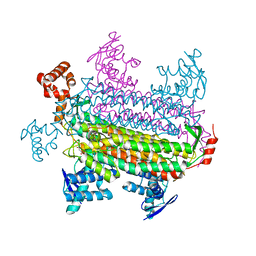 | |
3HGB
 
 | |
