5FKY
 
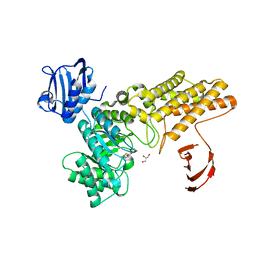 | | Structure of a hydrolase bound with an inhibitor | | Descriptor: | (3aR,5R,6S,7R,7aR)-2-amino-5-(hydroxymethyl)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, GLYCEROL, O-GLCNACASE BT_4395 | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
5FHG
 
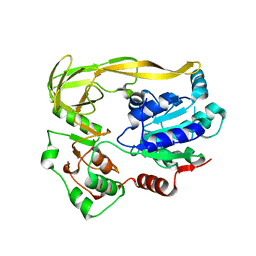 | |
5FHF
 
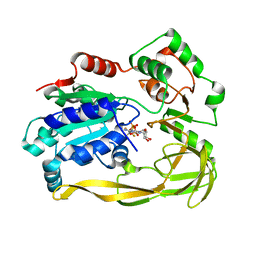 | | Crystal structure of Bacteroides sp Pif1 in complex with ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TETRAFLUOROALUMINATE ION, Uncharacterized protein | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5FHE
 
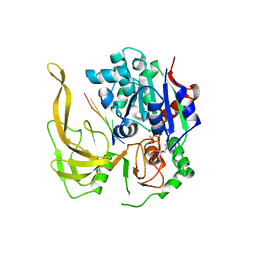 | | Crystal structure of Bacteroides Pif1 bound to ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5BWA
 
 | | Crystal structure of ODC-PLP-AZ1 ternary complex | | Descriptor: | Ornithine decarboxylase, Ornithine decarboxylase antizyme 1, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wu, D.H. | | Deposit date: | 2015-06-07 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of Ornithine Decarboxylase inactivation and accelerated degradation by polyamine sensor Antizyme1
Sci Rep, 5, 2015
|
|
5FL0
 
 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-(butylamino)-5-(hydroxymethyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d] [1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
5FHD
 
 | | Structure of Bacteroides sp Pif1 complexed with tailed dsDNA resulting in ssDNA bound complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*CP*CP*GP*GP*GP*GP*CP*CP*GP*CP*GP*C)-3'), MAGNESIUM ION, ... | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5FL1
 
 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-5-(hydroxymethyl)-2-(prop-2-enylamino)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
5FHH
 
 | | Structure of human Pif1 helicase domain residues 200-641 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, TETRAFLUOROALUMINATE ION | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5H48
 
 | | Crystal structure of Cbln1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5H4C
 
 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein Cbln4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
2Q6S
 
 | |
2Q6R
 
 | |
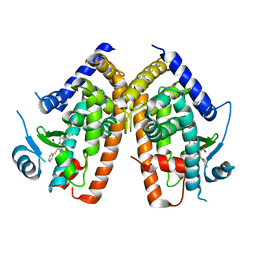
2Q5S
 
 | |
2Q61
 
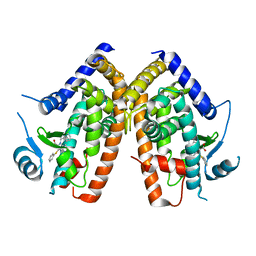 | |
5Z0W
 
 | |
6IWI
 
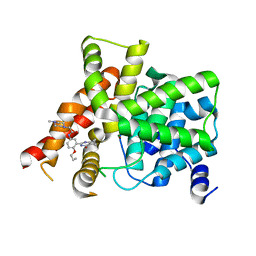 | | Crystal structure of PDE5A in complex with a novel inhibitor | | Descriptor: | MAGNESIUM ION, N-[3-(4,5-diethyl-6-oxo-1,6-dihydropyrimidin-2-yl)-4-propoxyphenyl]-2-(4-methylpiperazin-1-yl)acetamide, ZINC ION, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Pharmacokinetics-Driven Optimization of 4(3 H)-Pyrimidinones as Phosphodiesterase Type 5 Inhibitors Leading to TPN171, a Clinical Candidate for the Treatment of Pulmonary Arterial Hypertension.
J.Med.Chem., 62, 2019
|
|
6KTS
 
 | | Structure of C34N126K/N36 | | Descriptor: | Envelope glycoprotein, Glycoprotein 41 | | Authors: | Yu, D.W, Qin, B. | | Deposit date: | 2019-08-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Functional Characterization of the Secondary Mutation N126K Selected by Various HIV-1 Fusion Inhibitors.
Viruses, 12, 2020
|
|
2Q59
 
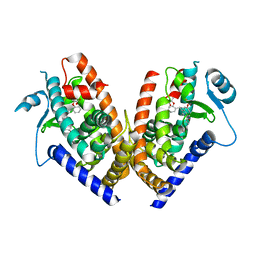 | | Crystal Structure of PPARgamma LBD bound to full agonist MRL20 | | Descriptor: | (2S)-2-(2-{[1-(4-METHOXYBENZOYL)-2-METHYL-5-(TRIFLUOROMETHOXY)-1H-INDOL-3-YL]METHYL}PHENOXY)PROPANOIC ACID, Peroxisome Proliferator-Activated Receptor gamma | | Authors: | Bruning, J.B, Nettles, K.W. | | Deposit date: | 2007-05-31 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Partial Agonists Activate PPARgamma Using a Helix 12 Independent Mechanism
Structure, 15, 2007
|
|
2Q5P
 
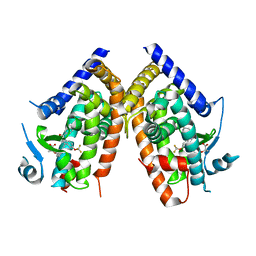 | | Crystal Structure of PPARgamma bound to partial agonist MRL24 | | Descriptor: | (2S)-2-(3-{[1-(4-METHOXYBENZOYL)-2-METHYL-5-(TRIFLUOROMETHOXY)-1H-INDOL-3-YL]METHYL}PHENOXY)PROPANOIC ACID, Peroxisome Proliferator-Activated Receptor gamma | | Authors: | Bruning, J.B, Nettles, K.W. | | Deposit date: | 2007-06-01 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Partial Agonists Activate PPARgamma Using a Helix 12 Independent Mechanism
Structure, 15, 2007
|
|
8RVF
 
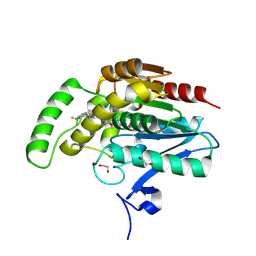 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 1,2-ETHANEDIOL, Monoglyceride lipase, [6-[[2,2-bis(fluoranyl)-1$l^{4},3-diaza-2$l^{4}-boratricyclo[7.3.0.0^{3,7}]dodeca-1(12),4,6,8,10-pentaen-8-yl]methyl]-2-azaspiro[3.3]heptan-2-yl]-[6-(3-cyclopropyl-1,2,4-triazol-1-yl)-2-azaspiro[3.3]heptan-2-yl]methanone | | Authors: | Leibrock, L, Hentsch, A, Nazare, M, Grether, U, Kuhn, B, Blaising, J, Benz, J. | | Deposit date: | 2024-02-01 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Highly Specific Miniaturized Fluorescent Monoacylglycerol Lipase Probes Enable Translational Research.
J.Am.Chem.Soc., 147, 2025
|
|
7VSQ
 
 | |
7VSP
 
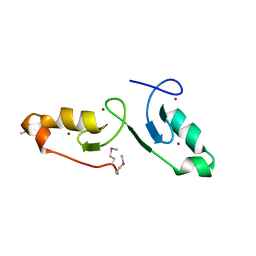 | | Crystal strcuture of the tandem B-Box domains of COL2 | | Descriptor: | ZINC ION, Zinc finger protein CONSTANS-LIKE 2 | | Authors: | Lv, X, Liu, R, Du, J. | | Deposit date: | 2021-10-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of CONSTANS oligomerization in FLOWERING LOCUS T activation.
J Integr Plant Biol, 64, 2022
|
|
8SDU
 
 | | Structure of rat organic anion transporter 1 (OAT1) | | Descriptor: | Solute carrier family 22 member 6 | | Authors: | Dou, T, Jiang, J. | | Deposit date: | 2023-04-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | The substrate and inhibitor binding mechanism of polyspecific transporter OAT1 revealed by high-resolution cryo-EM.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SDZ
 
 | |
