6E9F
 
 | | EsCas13d-crRNA-target RNA ternary complex | | Descriptor: | EsCas13d, MAGNESIUM ION, RNA (27-MER), ... | | Authors: | Zhang, C, Lyumkis, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d.
Cell, 175, 2018
|
|
3OUI
 
 | | PHD2-R717 with 40787422 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Egl nine homolog 1, ... | | Authors: | Arakaki, T.L, Kim, H. | | Deposit date: | 2010-09-14 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Benzimidazole-2-pyrazole HIF Prolyl 4-Hydroxylase Inhibitors as Oral Erythropoietin Secretagogues.
ACS Med Chem Lett, 1, 2010
|
|
8Z46
 
 | | SARS-CoV-2 3CL protease (3CL pro) in complex with a novel inhibitor | | Descriptor: | (2~{S})-~{N}-(3-azanyl-3-oxidanylidene-propyl)-4-[4-[[(1~{S})-1-(2-chlorophenyl)-3-oxidanyl-propyl]amino]-6-(methylamino)-1,3,5-triazin-2-yl]-1-ethanoyl-piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zhao, W.F, Xiong, L.W, Xu, Y.C. | | Deposit date: | 2024-04-16 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Covalent DNA-Encoded Library Workflow Drives Discovery of SARS-CoV-2 Nonstructural Protein Inhibitors.
J.Am.Chem.Soc., 146, 2024
|
|
8Z4W
 
 | | Crystal structures of SARS-CoV-2 papain-like protease in complex with covalent inhibitors | | Descriptor: | 1-[4-[[[4-(isoquinolin-5-ylamino)-6-(methylamino)-1,3,5-triazin-2-yl]amino]methyl]piperidin-1-yl]ethanone, GLYCEROL, Non-structural protein 3, ... | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Covalent DNA-Encoded Library Workflow Drives Discovery of SARS-CoV-2 Nonstructural Protein Inhibitors.
J.Am.Chem.Soc., 146, 2024
|
|
7X8R
 
 | | Cryo-EM structure of the Boc5-bound hGLP-1R-Gs complex | | Descriptor: | 2,4-bis(3-methoxy-4-thiophen-2-ylcarbonyloxy-phenyl)-1,3-bis[[4-[(2-methylpropan-2-yl)oxycarbonylamino]phenyl]carbonylamino]cyclobutane-1,3-dicarboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z.T, Zhou, Q.T, Li, Y, Chen, L.N, Zhang, Z.C, Liang, A.Y, Liu, Q, Wu, X.Y, Dai, A.T, Xia, T, Wu, W, Zhang, Y, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-03-14 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis of peptidomimetic agonism revealed by small- molecule GLP-1R agonists Boc5 and WB4-24.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X8S
 
 | | Cryo-EM structure of the WB4-24-bound hGLP-1R-Gs complex | | Descriptor: | 2,4-bis(3-methoxy-4-thiophen-2-ylcarbonyloxy-phenyl)-1,3-bis[[4-(2-methylpropanoylamino)phenyl]carbonylamino]cyclobutane-1,3-dicarboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z.T, Zhou, Q.T, Li, Y, Chen, L.N, Zhang, Z.C, Liang, A.Y, Liu, Q, Wu, X.Y, Dai, A.T, Xia, T, Wu, W, Zhang, Y, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-03-14 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of peptidomimetic agonism revealed by small- molecule GLP-1R agonists Boc5 and WB4-24.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2FZP
 
 | | Crystal structure of the USP8 interaction domain of human NRDP1 | | Descriptor: | ring finger protein 41 isoform 1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Amino-terminal Dimerization, NRDP1-Rhodanese Interaction, and Inhibited Catalytic Domain Conformation of the Ubiquitin-specific Protease 8 (USP8).
J.Biol.Chem., 281, 2006
|
|
3OUH
 
 | | PHD2-R127 with JNJ41536014 | | Descriptor: | 1-(5-chloro-6-fluoro-1H-benzimidazol-2-yl)-1H-pyrazole-4-carboxylic acid, Egl nine homolog 1, FE (II) ION, ... | | Authors: | Kim, H, Clark, R. | | Deposit date: | 2010-09-14 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Benzimidazole-2-pyrazole HIF Prolyl 4-Hydroxylase Inhibitors as Oral Erythropoietin Secretagogues.
ACS Med Chem Lett, 1, 2010
|
|
6BF4
 
 | |
3OUJ
 
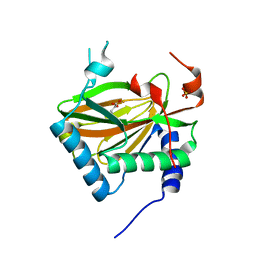 | | PHD2 with 2-Oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Egl nine homolog 1, FE (II) ION, ... | | Authors: | Staker, B.L, Arakaki, T.L. | | Deposit date: | 2010-09-14 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Benzimidazole-2-pyrazole HIF Prolyl 4-Hydroxylase Inhibitors as Oral Erythropoietin Secretagogues.
ACS Med Chem Lett, 1, 2010
|
|
3PIQ
 
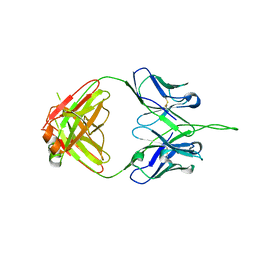 | | Crystal structure of human 2909 Fab, a quaternary structure-specific antibody against HIV-1 | | Descriptor: | Human monoclonal antibody 2909 Fab heavy chain, Human monoclonal antibody 2909 Fab light chain | | Authors: | Changela, A, Gorny, M.K, Zolla-Pazner, S, Kwong, P.D. | | Deposit date: | 2010-11-07 | | Release date: | 2011-01-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.325 Å) | | Cite: | Crystal Structure of Human Antibody 2909 Reveals Conserved Features of Quaternary Structure-Specific Antibodies That Potently Neutralize HIV-1.
J.Virol., 85, 2011
|
|
4E82
 
 | |
4E83
 
 | |
6IUP
 
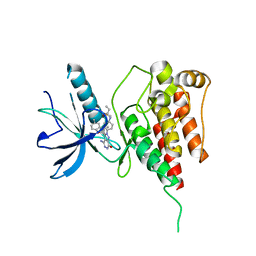 | | Crystal structure of FGFR4 kinase domain in complex with compound 5 | | Descriptor: | DIMETHYL SULFOXIDE, Fibroblast growth factor receptor 4, N-{4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6ITC
 
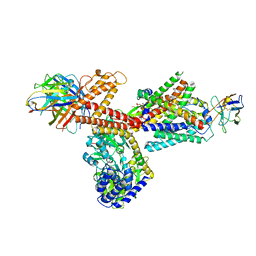 | | Structure of a substrate engaged SecA-SecY protein translocation machine | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ma, C.Y, Wu, X.F, Sun, D.J, Park, E.Y, Rapoport, T.A, Gao, N, Long, L. | | Deposit date: | 2018-11-21 | | Release date: | 2019-06-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the substrate-engaged SecA-SecY protein translocation machine.
Nat Commun, 10, 2019
|
|
6IUO
 
 | | Crystal structure of FGFR4 kinase domain in complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-({4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}methyl)prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6ITJ
 
 | | Crystal structure of FGFR1 kinase domain in complex with compound 3 | | Descriptor: | 4-azanyl-3-(3,5-dimethyl-1-benzofuran-2-yl)-2-phenyl-6~{H}-pyrazolo[3,4-d]pyridazin-7-one, Fibroblast growth factor receptor 1 | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-23 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
9HVW
 
 | |
9KNY
 
 | | Cryo-EM structure of pyruvate-treated human mitochondrial pyruvate carrier in the IMS-open conformation at pH 8.0 | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, MPC specific nanobody 1, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-11-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
9KNX
 
 | | Cryo-EM structure of human mitochondrial pyruvate carrier in the occluded conformation at pH 6.8 | | Descriptor: | CARDIOLIPIN, MPC specific nanobody 1, Mitochondrial pyruvate carrier 1, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-11-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
9KNW
 
 | | Cryo-EM structure of apo human mitochondrial pyruvate carrier in the IMS-open conformation at pH 6.8 | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, MPC specific nanobody 1, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-11-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
4HPZ
 
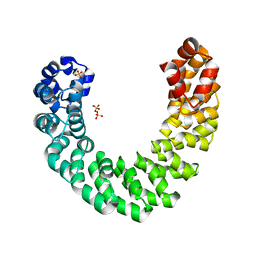 | |
8UC8
 
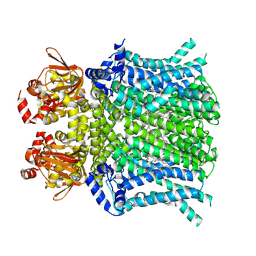 | | HCN1 nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Kim, E.D, Nimigean, C.M. | | Deposit date: | 2023-09-25 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Propofol rescues voltage-dependent gating of HCN1 channel epilepsy mutants.
Nature, 632, 2024
|
|
8UC7
 
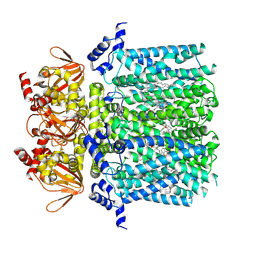 | | HCN1 complex with propofol | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,6-BIS(1-METHYLETHYL)PHENOL, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Kim, E.D, Nimigean, C.M. | | Deposit date: | 2023-09-25 | | Release date: | 2024-07-31 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Propofol rescues voltage-dependent gating of HCN1 channel epilepsy mutants.
Nature, 632, 2024
|
|
4QBS
 
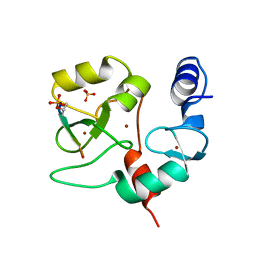 | |
