7XYR
 
 | |
2LZ4
 
 | |
7CDB
 
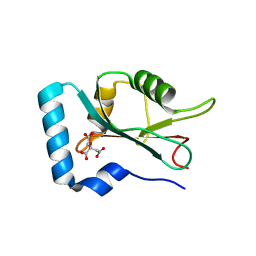 | | Structure of GABARAPL1 in complex with GABA(A) receptor gamma 2 | | Descriptor: | ACETATE ION, CITRIC ACID, Gamma-aminobutyric acid receptor subunit gamma-2, ... | | Authors: | Li, J, Ye, J, Zhu, R, Kong, C, Zhang, M, Wang, C. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural basis of GABARAP-mediated GABA A receptor trafficking and functions on GABAergic synaptic transmission.
Nat Commun, 12, 2021
|
|
8XOQ
 
 | | Human Calcium and Integrin Binding Protein 2 (CIB2) Fusion to TMC1 CBD-1 domain at 2.4 Angstroms resolution | | Descriptor: | MAGNESIUM ION, Transmembrane channel-like protein 1,Calcium and integrin-binding family member 2 | | Authors: | Li, Y.H, Chen, J.S, Zhang, X, Wang, C. | | Deposit date: | 2024-01-02 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural insights into calcium-dependent CIB2-TMC1 interaction in hair cell mechanotransduction.
Commun Biol, 8, 2025
|
|
7DHS
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1R)-1-phenylethyl]benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Wu, T, Xiang, Q, Wang, C, Wu, C, Zhang, C, Zhang, M, Liu, Z, Zhang, Y, Xiao, L, Xu, Y. | | Deposit date: | 2020-11-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Y06014 is a selective BET inhibitor for the treatment of prostate cancer.
Acta Pharmacol.Sin., 42, 2021
|
|
2MY5
 
 | | Solution Structure of KstB-PCP in kosinostatin biosynthesis | | Descriptor: | N~3~-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-(2-sulfanylethyl)-beta-alaninamide, Peptidyl carrier protein | | Authors: | Zhao, B, Lan, W, Wang, C, Tang, G, Cao, C. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N Assigned Chemical Shifts for KstB-PCP
To be Published
|
|
2MY6
 
 | |
2MVT
 
 | | Solution structure of scoloptoxin SSD609 from Scolopendra mutilans | | Descriptor: | Scoloptoxin SSD609 | | Authors: | Wu, F, Sun, P, Wang, C, He, Y, Zhang, L, Tian, C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A distinct three-helix centipede toxin SSD609 inhibits Iks channels by interacting with the KCNE1 auxiliary subunit.
Sci Rep, 5, 2015
|
|
6JHZ
 
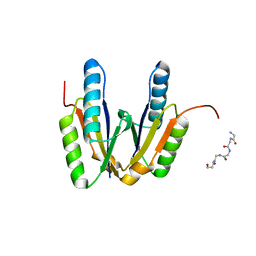 | | Crystal structure of cas2 | | Descriptor: | 5-mer peptide, CRISPR-associated endoribonuclease Cas2 | | Authors: | Bi, M, Mo, X, Wang, C, Yuan, A.Y. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural insight into endonuclease CRISPR-associated Cas2 protein
To be published
|
|
6IPB
 
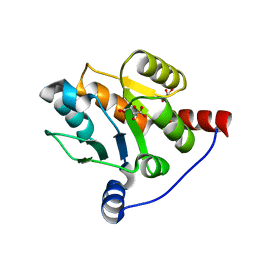 | |
6IZJ
 
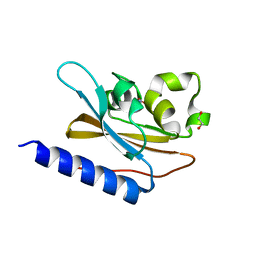 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, NreA | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2018-12-19 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
6K2H
 
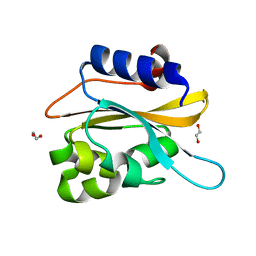 | | structural characterization of mutated NreA protein in nitrate binding site from staphylococcus aureus. | | Descriptor: | 1,2-ETHANEDIOL, NreA | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2019-05-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
6L94
 
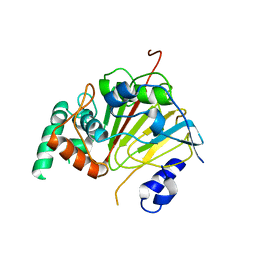 | | The structure of the dioxygenase ABH1 from mouse | | Descriptor: | FE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Xie, W, Wang, C, Li, H, Zhang, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.10012341 Å) | | Cite: | ALKBH1 promotes lung cancer by regulating m6A RNA demethylation.
Biochem Pharmacol, 189, 2021
|
|
7VYQ
 
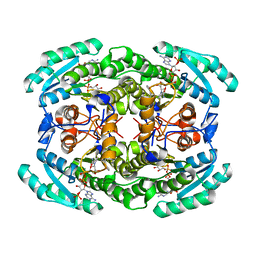 | | Short chain dehydrogenase (SCR) cryoEM structure with NADP and ethyl 4-chloroacetoacetate | | Descriptor: | Carbonyl Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ethyl 4-chloranyl-3-oxidanylidene-butanoate | | Authors: | Li, Y.H, Zhang, R.Z, Wang, C, Forouhar, F, Clarke, O, Vorobiev, S, Singh, S, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2021-11-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7XCE
 
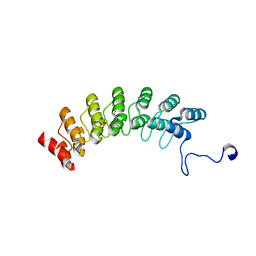 | | Crystal structure of Ankyrin G in complex with neurofascin | | Descriptor: | GLYCEROL, Neurofascin,Ankyrin-3, SULFATE ION | | Authors: | He, L, Li, J, Wang, C. | | Deposit date: | 2022-03-24 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Ankyrin-G in complex with a fragment of Neurofascin reveals binding mechanisms required for integrity of the axon initial segment.
J.Biol.Chem., 298, 2022
|
|
7XNG
 
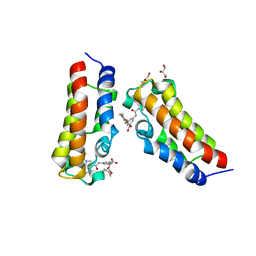 | | Crystal structure of CBP bromodomain liganded with Y08092(31g) | | Descriptor: | 3-[(1-ethanoylindol-3-yl)carbonylamino]-5-[[(2S)-oxan-2-yl]oxymethyl]benzoic acid, CREB-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M, Xu, Y. | | Deposit date: | 2022-04-28 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of CBP bromodomain liganded with Y08092(31g)
To Be Published
|
|
7XM7
 
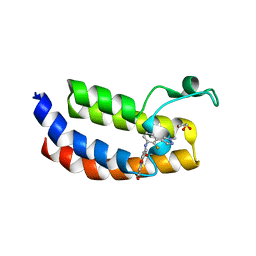 | | Crystal Structure of the CBP in complex with the Y08188 | | Descriptor: | 1,2-ETHANEDIOL, 3-ethanoyl-~{N}-[2-fluoranyl-3-(1-methylpyrazol-4-yl)phenyl]-7-methoxy-indolizine-1-carboxamide, CREB-binding protein, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M, Xu, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as potent and selective CBP bromodomain inhibitors
To Be Published
|
|
8Y2L
 
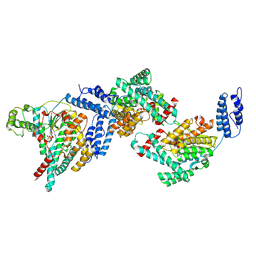 | | The Crystal Structure of Glucosyltransferase TcdB from Clostridioides difficile | | Descriptor: | Glucosyltransferase TcdB, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Fan, S, Wei, X, Lv, R, Wang, C, Tang, M, Jin, Y, Yang, Z. | | Deposit date: | 2024-01-26 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | The Crystal Structure of Glucosyltransferase TcdB from Clostridioides difficile
To Be Published
|
|
8XND
 
 | | Crystal structure of serine hydroxymethyltransferase 1 | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, cytosolic | | Authors: | Fan, S, Wei, X, Lv, R, Wang, C, Tang, M, Jin, Y, Yang, Z. | | Deposit date: | 2023-12-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structure of serine hydroxymethyltransferase 1
To Be Published
|
|
8XNA
 
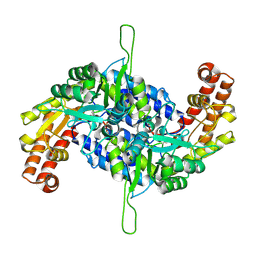 | | Crystal structure of serine hydroxymethyltransferase 2 | | Descriptor: | Serine hydroxymethyltransferase, mitochondrial | | Authors: | Fan, S, Lv, R, Wang, C, Tang, M, Wei, X, Jin, Y, Yang, Z. | | Deposit date: | 2023-12-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of serine hydroxymethyltransferase 2
To Be Published
|
|
2K7K
 
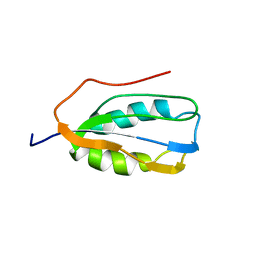 | | Human Acylphosphatase (AcPh) common type | | Descriptor: | Acylphosphatase-1 | | Authors: | Gribenko, A.V, Patel, M.M, Liu, J, McCallum, S.A, Wang, C, Makhatadze, G.I. | | Deposit date: | 2008-08-13 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Rational stabilization of enzymes by computational redesign of surface charge-charge interactions
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2K7J
 
 | | Human Acylphosphatase(AcPh) surface charge-optimized | | Descriptor: | Acylphosphatase-1 | | Authors: | Gribenko, A.V, Patel, M.M, Liu, J, McCallum, S.A, Wang, C, Makhatadze, G.I. | | Deposit date: | 2008-08-12 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Rational stabilization of enzymes by computational redesign of surface charge-charge interactions
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2L8L
 
 | |
7DMG
 
 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADP | | Descriptor: | (S)-specific carbonyl reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-12-03 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLM
 
 | | Short chain dehydrogenase (SCR) crystal structure with NADPH | | Descriptor: | Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-28 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
