3C0R
 
 | |
3LRS
 
 | | Structure of PG16, an antibody with broad and potent neutralization of HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PG-16 Heavy Chain Fab, PG-16 Light Chain Fab | | Authors: | Pancera, M, Kwong, P.D. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1.
J.Virol., 84, 2010
|
|
2ZDZ
 
 | | X-ray structure of Bace-1 in complex with compound 3.b.10 | | Descriptor: | Beta-secretase 1, N-carbamimidoyl-2-[2-(2-chlorophenyl)-5-[4-(4-ethanoylphenoxy)phenyl]pyrrol-1-yl]ethanamide | | Authors: | Chopra, R, Olland, A. | | Deposit date: | 2007-12-04 | | Release date: | 2008-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acylguanidine inhibitors of beta-secretase: optimization of the pyrrole ring substituents extending into the S1 and S3 substrate binding pockets.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3KUF
 
 | | The Crystal Structure of the Tudor Domains from FXR1 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Fragile X mental retardation syndrome-related protein 1, GLYCEROL | | Authors: | Bian, C, Guo, Y.H, Adams-Cioaba, M.A, Mackenzie, F, Kozieradzki, I, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-27 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Tudor domains from Fragile X mental retardation syndrome-related protein 1
To be Published
|
|
3BW1
 
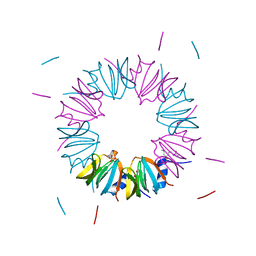 | | Crystal structure of homomeric yeast Lsm3 exhibiting novel octameric ring organisation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, SULFATE ION, U6 snRNA-associated Sm-like protein LSm3 | | Authors: | Naidoo, N, Harrop, S.J, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2008-01-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Lsm3 octamer from Saccharomyces cerevisiae: implications for Lsm ring organisation and recruitment
J.Mol.Biol., 377, 2008
|
|
3HYC
 
 | |
3BRK
 
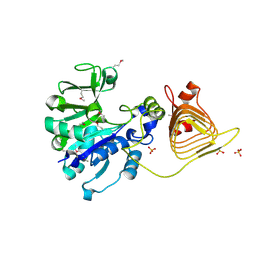 | |
3LOK
 
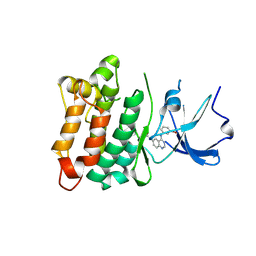 | |
3KE1
 
 | |
3L08
 
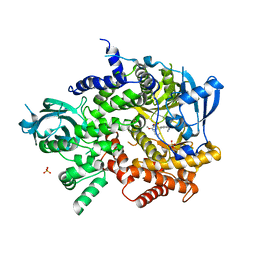 | | Structure of Pi3K gamma with a potent inhibitor: GSK2126458 | | Descriptor: | 2,4-difluoro-N-[2-methoxy-5-(4-pyridazin-4-ylquinolin-6-yl)pyridin-3-yl]benzenesulfonamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Elkins, P.A, Marrero, E.M. | | Deposit date: | 2009-12-09 | | Release date: | 2010-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of GSK2126458, a Highly Potent Inhibitor of PI3K and the Mammalian Target of Rapamycin.
ACS Med Chem Lett, 1, 2010
|
|
3KG9
 
 | |
3KG7
 
 | |
3CD2
 
 | | LIGAND INDUCED CONFORMATIONAL CHANGES IN THE CRYSTAL STRUCTURES OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE COMPLEXES WITH FOLATE AND NADP+ | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Rak, D, Luft, J, Pangborn, W, Queener, S. | | Deposit date: | 1999-03-16 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-induced conformational changes in the crystal structures of Pneumocystis carinii dihydrofolate reductase complexes with folate and NADP+.
Biochemistry, 38, 1999
|
|
3CAL
 
 | |
3L14
 
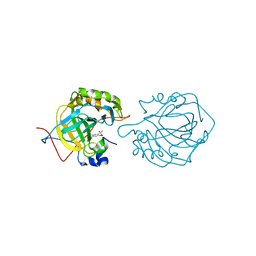 | | Human Carbonic Anhydrase II complexed with Althiazide | | Descriptor: | 4-AMINO-6-CHLOROBENZENE-1,3-DISULFONAMIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Quirit, J.G, Robbins, A, Genis, C, Tu, C, Silverman, D.N, McKenna, R. | | Deposit date: | 2009-12-10 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | The Promiscuous Nature of Althiazide in Adducts with CA II and CA IX Mimic
To be Published
|
|
2ZE1
 
 | | X-ray structure of Bace-1 in complex with compound 6g | | Descriptor: | 3-bromo-N-[4-[1-(2-carbamimidamido-2-oxo-ethyl)-5-phenyl-pyrrol-2-yl]phenyl]benzamide, Beta-secretase 1 | | Authors: | Chopra, R, Olland, A. | | Deposit date: | 2007-12-05 | | Release date: | 2008-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acylguanidine inhibitors of beta-secretase: optimization of the pyrrole ring substituents extending into the S1 and S3 substrate binding pockets.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3LJT
 
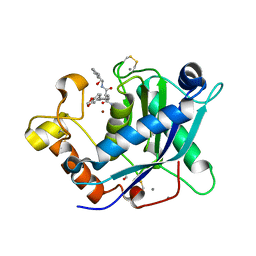 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with an Amino-2-indanol compound | | Descriptor: | (2R)-2-[4-(1,3-benzodioxol-5-yl)benzyl]-N~4~-hydroxy-N~1~-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]butanediamide, 1,2-ETHANEDIOL, A disintegrin and metalloproteinase with thrombospondin motifs 5, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N. | | Deposit date: | 2010-01-26 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure analysis reveals the flexibility of the ADAMTS-5 active site.
Protein Sci., 20, 2011
|
|
3CJF
 
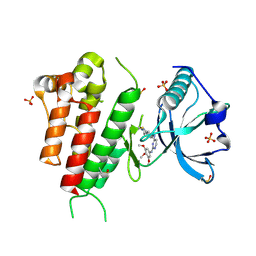 | | Crystal structure of VEGFR2 in complex with a 3,4,5-trimethoxy aniline containing pyrimidine | | Descriptor: | N~4~-(3-methyl-1H-indazol-6-yl)-N~2~-(3,4,5-trimethoxyphenyl)pyrimidine-2,4-diamine, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Nolte, R.T. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of 5-[[4-[(2,3-dimethyl-2H-indazol-6-yl)methylamino]-2-pyrimidinyl]amino]-2-methyl-benzenesulfonamide (Pazopanib), a novel and potent vascular endothelial growth factor receptor inhibitor.
J.Med.Chem., 51, 2008
|
|
3CEN
 
 | |
3CKD
 
 | | Crystal structure of the C-terminal domain of the Shigella type III effector IpaH | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Invasion plasmid antigen, ... | | Authors: | Lam, R, Singer, A.U, Cuff, M.E, Skarina, T, Kagan, O, DiLeo, R, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3CJG
 
 | | Crystal structure of VEGFR2 in complex with a 3,4,5-trimethoxy aniline containing pyrimidine | | Descriptor: | N~4~-methyl-N~4~-(3-methyl-1H-indazol-6-yl)-N~2~-(3,4,5-trimethoxyphenyl)pyrimidine-2,4-diamine, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Nolte, R.T. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of 5-[[4-[(2,3-dimethyl-2H-indazol-6-yl)methylamino]-2-pyrimidinyl]amino]-2-methyl-benzenesulfonamide (Pazopanib), a novel and potent vascular endothelial growth factor receptor inhibitor.
J.Med.Chem., 51, 2008
|
|
3D5J
 
 | | Structure of yeast Grx2-C30S mutant with glutathionyl mixed disulfide | | Descriptor: | GLUTATHIONE, Glutaredoxin-2, mitochondrial | | Authors: | Discola, K.F, de Oliveira, M.A, Barcena, J.A, Porras, P, Padilla, C.A, Guimaraes, B.G, Netto, L.E.S. | | Deposit date: | 2008-05-16 | | Release date: | 2008-10-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural aspects of the distinct biochemical properties of glutaredoxin 1 and glutaredoxin 2 from Saccharomyces cerevisiae.
J.Mol.Biol., 385, 2009
|
|
3MME
 
 | | Structure and functional dissection of PG16, an antibody with broad and potent neutralization of HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG16 HEAVY CHAIN FAB, ... | | Authors: | Pancera, M, McLellan, J, Zhou, T, Zhu, J, Kwong, P. | | Deposit date: | 2010-04-19 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.97 Å) | | Cite: | Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1.
J.Virol., 84, 2010
|
|
4E0V
 
 | | Structure of L-amino acid oxidase from the B. jararacussu venom | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-amino-acid oxidase | | Authors: | Ullah, A, Souza, T.A.C.B, Betzel, C, Murakami, M.T, Arni, R.K. | | Deposit date: | 2012-03-05 | | Release date: | 2012-04-25 | | Last modified: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into selectivity and cofactor binding in snake venom L-amino acid oxidases.
Biochem.Biophys.Res.Commun., 421, 2012
|
|
3NQS
 
 | | Crystal Structure of Inducible Nitric Oxide Synthase with N-Nitrosated-pterin | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYL 4-[(4-METHYLPYRIDIN-2-YL)AMINO]PIPERIDINE-1-CARBOXYLATE, ... | | Authors: | Rosenfeld, R.J, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2010-06-29 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nitric-oxide synthase forms N-NO-pterin and S-NO-cys: implications for activity, allostery, and regulation.
J.Biol.Chem., 285, 2010
|
|
