6SMG
 
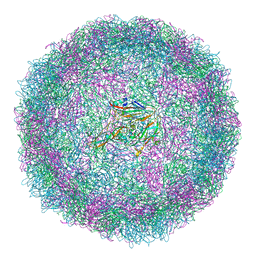 | | Structure of Coxsackievirus A10 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Zhao, Y, Zhou, D, Ni, T, Karia, D, Kotecha, A, Wang, X, Rao, Z, Jones, E.Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2019-08-21 | | Release date: | 2020-01-15 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10.
Nat Commun, 11, 2020
|
|
3I6Z
 
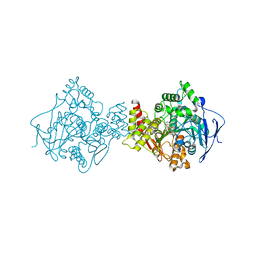 | | 3D Structure of Torpedo californica acetylcholinesterase complexed with N-saccharinohexyl-galanthamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{6-[(4aS,6R,8aS)-6-hydroxy-3-methoxy-5,6,9,10-tetrahydro-4aH-[1]benzofuro[3a,3,2-ef][2]benzazepin-11(12H)-yl]hexyl}-1 ,2-benzisothiazol-3(2H)-one 1,1-dioxide, ... | | Authors: | Lamba, D, Bartolucci, C. | | Deposit date: | 2009-07-07 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Probing Torpedo californica acetylcholinesterase catalytic gorge with two novel bis-functional galanthamine derivatives.
J.Med.Chem., 53, 2010
|
|
2XQB
 
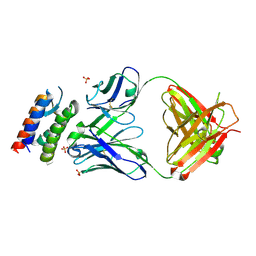 | | Crystal Structure of anti-IL-15 Antibody in Complex with human IL-15 | | Descriptor: | ANTI-IL-15 ANTIBODY, INTERLEUKIN 15, SULFATE ION | | Authors: | Lowe, D.C, Gerhardt, S, Ward, A, Hargreaves, D, Anderson, M, StGallay, S, Vousden, K, Ferraro, F, Pauptit, R.A, Cochrane, D, Pattison, D.V, Buchanan, C, Popovic, B, Finch, D.K, Wilkinson, T, Sleeman, M, Vaughan, T.J, Cruwys, S, Mallinder, P.R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a High Affinity Anti-Il-15 Antibody: Crystal Structure Reveals an Alpha-Helix in Vh Cdr3 as Key Component of Paratope.
J.Mol.Biol., 406, 2011
|
|
2FBW
 
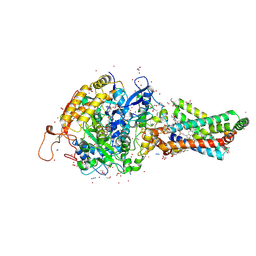 | | Avian respiratory complex II with carboxin bound | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, AZIDE ION, ... | | Authors: | Huang, L.S, Sun, G, Cobessi, D, Wang, A.C, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-12-10 | | Release date: | 2005-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme.
J.Biol.Chem., 281, 2006
|
|
7BBD
 
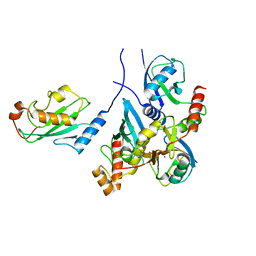 | | Crystal structure of monoubiquitinated TRIM21 RING (Ub-RING) In complex with ubiquitin charged Ube2N (Ube2N~Ub) and Ube2V2 | | Descriptor: | Polyubiquitin-B,E3 ubiquitin-protein ligase TRIM21, Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Kiss, L, Neuhaus, D, James, L.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RING domains act as both substrate and enzyme in a catalytic arrangement to drive self-anchored ubiquitination.
Nat Commun, 12, 2021
|
|
7ZQ7
 
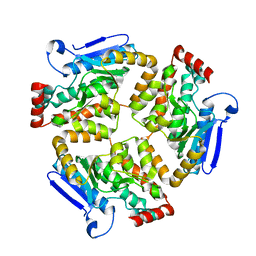 | | Structure of RpF-1 | | Descriptor: | Crotonase/enoyl-CoA hydratase family protein | | Authors: | Sanchez-Alba, L, Reverter, D, Conchillo, O, Yero, D, Daura, X, Gibert, I. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of RpF-1
To Be Published
|
|
3GGO
 
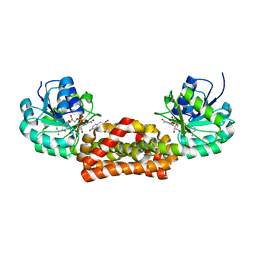 | | Crystal structure of prephenate dehydrogenase from A. aeolicus with HPP and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Prephenate dehydrogenase | | Authors: | Sun, W, Shahinas, D, Christendat, D. | | Deposit date: | 2009-03-01 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Aquifex aeolicus Prephenate Dehydrogenase Reveals the Mode of Tyrosine Inhibition.
J.Biol.Chem., 284, 2009
|
|
2C5B
 
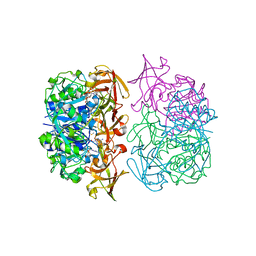 | | X-ray crystal structure of 5'-fluorodeoxyadenosine synthase from Streptomyces cattleya complexed with 2'deoxy-5'deoxy-fluoroadenosine. | | Descriptor: | 5'-FLUORO-2',5'-DIDEOXYADENOSINE, 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, METHIONINE | | Authors: | McEwan, A.R, Deng, H, McGlinchey, R.P, Robinson, D.A, O'Hagan, D, Naismith, J.H, Spencer, J. | | Deposit date: | 2005-10-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Specificity in Enzymatic Fluorination. The Fluorinase from Streptomyces Cattleya Accepts 2'-Deoxyadenosine Substrates.
Org.Biomol.Chem., 4, 2006
|
|
5ABY
 
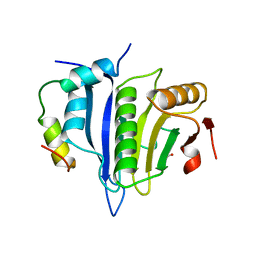 | | Complex of C. elegans eIF4E-3 with the 4E-binding protein Mextli | | Descriptor: | 4E-BINDING PROTEIN MEXTLI, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-3, GLYCEROL, ... | | Authors: | Peter, D, Weichenrieder, O. | | Deposit date: | 2015-08-09 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mextli Proteins Use Both Canonical Bipartite and Novel Tripartite Binding Modes to Form Eif4E Complexes that Display Differential Sensitivity to 4E-BP Regulation
Genes Dev., 29, 2015
|
|
2BRZ
 
 | | SOLUTION NMR STRUCTURE OF THE SWEET PROTEIN BRAZZEIN, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BRAZZEIN | | Authors: | Caldwell, J.E, Abildgaard, F, Dzakula, Z, Ming, D, Hellekant, G, Markley, J.L. | | Deposit date: | 1998-04-30 | | Release date: | 1998-07-01 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the thermostable sweet-tasting protein brazzein.
Nat.Struct.Biol., 5, 1998
|
|
2X8H
 
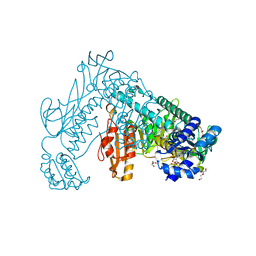 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with GSH | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Angelucci, F, Dimastrogiovanni, D, Boumis, G, Brunori, M, Miele, A.E, Saccoccia, F, Bellelli, A. | | Deposit date: | 2010-03-09 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mapping the Catalytic Cycle of Schistosoma Mansoni Thioredoxin Glutathione Reductase by X-Ray Crystallography
J.Biol.Chem., 285, 2010
|
|
1ZM7
 
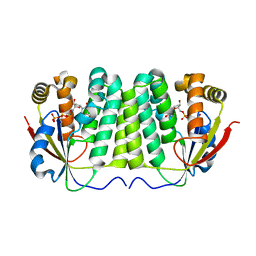 | | Crystal structure of D. melanogaster deoxyribonucleoside kinase mutant N64D in complex with dTTP | | Descriptor: | Deoxynucleoside kinase, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Welin, M, Skovgaard, T, Knecht, W, Berenstein, D, Munch-Petersen, B, Piskur, J, Eklund, H. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the changed substrate specificity of Drosophila melanogaster deoxyribonucleoside kinase mutant N64D.
Febs J., 272, 2005
|
|
2IQ7
 
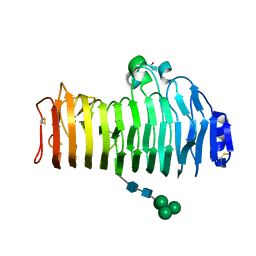 | | Crystal structure of the polygalacturonase from Colletotrichum lupini and its implications for the interaction with polygalacturonase-inhibiting proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bonivento, D, Federici, L, Matteo, A.D. | | Deposit date: | 2006-10-13 | | Release date: | 2007-10-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the endopolygalacturonase from the phytopathogenic fungus Colletotrichum lupini and its interaction with polygalacturonase-inhibiting proteins
Proteins, 70, 2008
|
|
3I6M
 
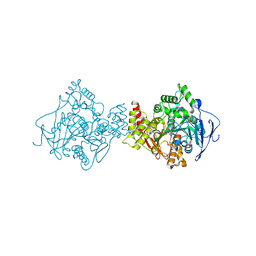 | | 3D Structure of Torpedo californica acetylcholinesterase complexed with N-piperidinopropyl-galanthamine | | Descriptor: | (4aS,6R,8aS)-3-methoxy-11-(3-piperidin-1-ylpropyl)-5,6,9,10,11,12-hexahydro-4aH-[1]benzofuro[3a,3,2-ef][2]benzazepin-6-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lamba, D, Bartolucci, C. | | Deposit date: | 2009-07-07 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Probing Torpedo californica acetylcholinesterase catalytic gorge with two novel bis-functional galanthamine derivatives.
J.Med.Chem., 53, 2010
|
|
1O6Z
 
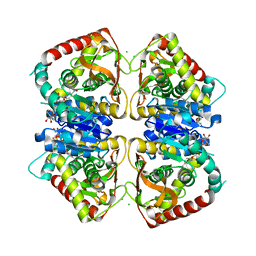 | | 1.95 A resolution structure of (R207S,R292S) mutant of malate dehydrogenase from the halophilic archaeon Haloarcula marismortui (holo form) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Irimia, A, Ebel, C, Madern, D, Richard, S.B, Cosenza, L.W, Zaccai, G, Vellieux, F.M.D. | | Deposit date: | 2002-10-22 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Oligomeric States of Haloarcula Marismortui Malate Dehydrogenase are Modulated by Solvent Components as Shown by Crystallographic and Biochemical Studies
J.Mol.Biol., 326, 2003
|
|
2VTT
 
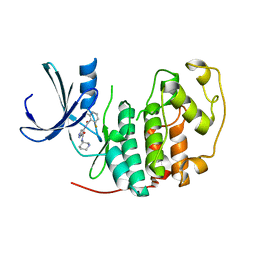 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | 4-{[(2,6-difluorophenyl)carbonyl]amino}-N-[(3S)-piperidin-3-yl]-1H-pyrazole-3-carboxamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
6SNB
 
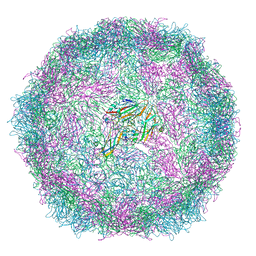 | | Structure of Coxsackievirus A10 A-particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Zhao, Y, Zhou, D, Ni, T, Karia, D, Kotecha, A, Wang, X, Rao, Z, Jones, E.Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2019-08-23 | | Release date: | 2020-01-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10.
Nat Commun, 11, 2020
|
|
6DXK
 
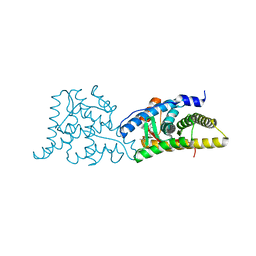 | | Glucocorticoid Receptor in complex with Compound 11 | | Descriptor: | (8S,11R,13S,14S,17S)-11-[4-(dimethylamino)phenyl]-17-(3,3-dimethylbut-1-yn-1-yl)-17-hydroxy-13-methyl-1,2,6,7,8,11,12,13,14,15,16,17-dodecahydro-3H-cyclopenta[a]phenanthren-3-one (non-preferred name), Glucocorticoid receptor | | Authors: | Rew, Y, Du, X, Eksterowicz, J, Zhou, H, Jahchan, N, Zhu, L, Yan, X, Kawai, H, McGee, L.R, Medina, J.C, Huang, T, Chen, C, Zavorotinskaya, T, Sutimantanapi, D, Waszczuk, J, Jackson, E, Huang, E, Ye, Q, Fantin, V.R, Daqing, S. | | Deposit date: | 2018-06-29 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of a Potent and Selective Steroidal Glucocorticoid Receptor Antagonist (ORIC-101).
J. Med. Chem., 61, 2018
|
|
7ND3
 
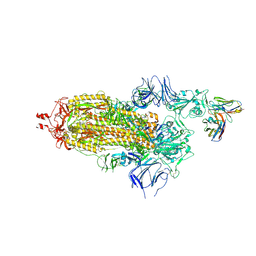 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-40 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-40 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND4
 
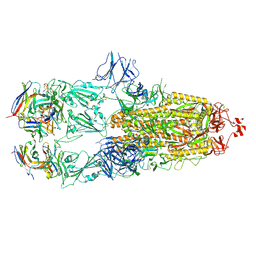 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-88 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-88 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDC
 
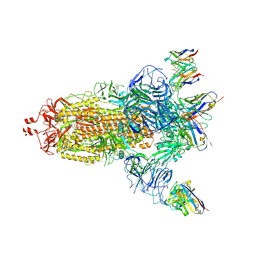 | | EM structure of SARS-CoV-2 Spike glycoprotein (all RBD down) in complex with COVOX-159 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-159 Fab light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND7
 
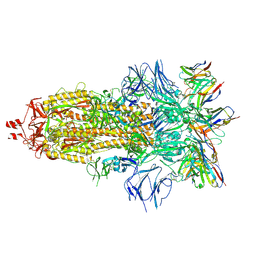 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-316 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND9
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein (one RBD up) in complex with COVOX-253H55L Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDA
 
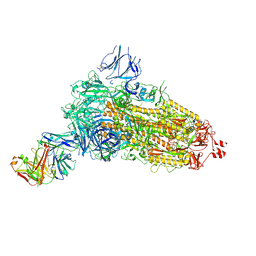 | | EM structure of SARS-CoV-2 Spike glycoprotein (all RBD down) in complex with COVOX-253H55L Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
2VTH
 
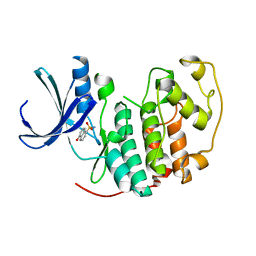 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H-pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | Descriptor: | 5-hydroxynaphthalene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2, GLYCEROL | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
