7DWA
 
 | | Structure of a novel beta-mannanase BaMan113A with mannotriose, N236Y mutation | | Descriptor: | Endo-beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Liu, W.T, Liu, W.D, Zheng, Y.Y. | | Deposit date: | 2021-01-15 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Functional and structural investigation of a novel beta-mannanase BaMan113A from Bacillus sp. N16-5.
Int.J.Biol.Macromol., 182, 2021
|
|
7DW8
 
 | | Structure of a novel beta-mannanase BaMan113A with mannobiose, N236Y mutation. | | Descriptor: | Endo-beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Liu, W.T, Liu, W.D, Zheng, Y.Y. | | Deposit date: | 2021-01-15 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural investigation of a novel beta-mannanase BaMan113A from Bacillus sp. N16-5.
Int.J.Biol.Macromol., 182, 2021
|
|
7DV7
 
 | |
7DVJ
 
 | | Structure of beta-mannanase BaMan113A with mannobiose | | Descriptor: | Endo-beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Liu, W.T, Liu, W.D, Zheng, Y.Y. | | Deposit date: | 2021-01-13 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and structural investigation of a novel beta-mannanase BaMan113A from Bacillus sp. N16-5.
Int.J.Biol.Macromol., 182, 2021
|
|
7DVZ
 
 | |
7DEK
 
 | | Pseudomonas aeruginosa FK506-binding protein PaFkbA | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Ynag, J, Huang, Q, Bao, R. | | Deposit date: | 2020-11-04 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization of PaFkbA: A periplasmic chaperone from Pseudomonas aeruginosa .
Comput Struct Biotechnol J, 19, 2021
|
|
4OYD
 
 | | Crystal structure of a computationally designed inhibitor of an Epstein-Barr viral Bcl-2 protein | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BHRF1, Computationally designed Inhibitor | | Authors: | Shen, B, Procko, E, Baker, D, Stoddard, B. | | Deposit date: | 2014-02-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A computationally designed inhibitor of an epstein-barr viral bcl-2 protein induces apoptosis in infected cells.
Cell, 157, 2014
|
|
6AK2
 
 | | Crystal structure of the syntenin PDZ1 domain in complex with the peptide inhibitor KSL-128018 | | Descriptor: | Syntenin-1, peptide inhibitor KSL-128018 | | Authors: | Jin, Z.Y, Park, J.H, Yun, J.H, Haugaard-Kedstrom, L.M, Lee, W.T. | | Deposit date: | 2018-08-29 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | A High-Affinity Peptide Ligand Targeting Syntenin Inhibits Glioblastoma.
J.Med.Chem., 64, 2021
|
|
4Q2O
 
 | |
4Q2Q
 
 | |
4Q2P
 
 | | NHERF3 PDZ2 in Complex with a Phage-Derived Peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Na(+)/H(+) exchange regulatory cofactor NHE-RF3 | | Authors: | Appleton, B.A, Wiesmann, C. | | Deposit date: | 2014-04-09 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural portrait of the PDZ domain family.
J.Mol.Biol., 426, 2014
|
|
4Q2N
 
 | |
2ZCS
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with bisphosphonate BPH-700 | | Descriptor: | Dehydrosqualene synthase, tripotassium (1R)-4-biphenyl-4-yl-1-phosphonatobutane-1-sulfonate | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H, Oldfield, E. | | Deposit date: | 2007-11-11 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence.
Science, 319, 2008
|
|
2HT6
 
 | |
5T5S
 
 | | A fragment of a human tRNA synthetase | | Descriptor: | Alanine--tRNA ligase, cytoplasmic | | Authors: | Sun, L, Schimmel, P. | | Deposit date: | 2016-08-31 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Two crystal structures reveal design for repurposing the C-Ala domain of human AlaRS.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5T76
 
 | | A fragment of a human tRNA synthetase | | Descriptor: | Alanine--tRNA ligase, cytoplasmic | | Authors: | Sun, L, Schimmel, P. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two crystal structures reveal design for repurposing the C-Ala domain of human AlaRS.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5SBF
 
 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 NendoU | | Descriptor: | CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-07-23 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
4MV5
 
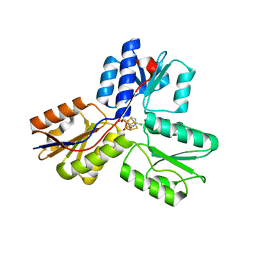 | | IspH in complex with 6-chloropyridin-3-ylmethyl diphosphate | | Descriptor: | (6-chloropyridin-3-yl)methyl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
5S72
 
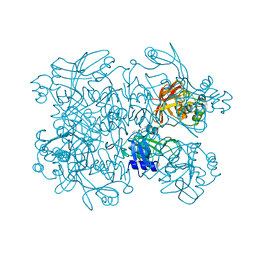 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with BBL029427 | | Descriptor: | CITRIC ACID, N-(2-aminoethyl)-N'-phenylurea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.512 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S6Z
 
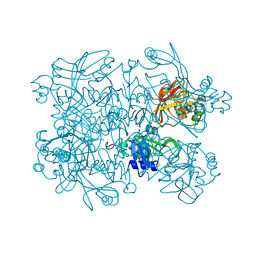 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with PB2255187532 | | Descriptor: | 4-[(dimethylamino)methyl]-1,3-thiazol-2-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S6Y
 
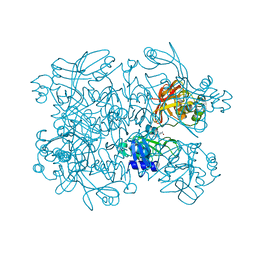 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z56900771 | | Descriptor: | CITRIC ACID, N-[(furan-2-yl)methyl]urea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S71
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with FUZS-5 | | Descriptor: | 5'-thiothymidine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Bege, M, Bajusz, D, BorbAs, A, Keseru, G.M, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
2ZCQ
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with bisphosphonate BPH-652 | | Descriptor: | (1R)-4-(3-phenoxyphenyl)-1-phosphonobutane-1-sulfonic acid, Dehydrosqualene synthase, MAGNESIUM ION | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H, Oldfield, E. | | Deposit date: | 2007-11-11 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence.
Science, 319, 2008
|
|
2ZCO
 
 | |
2ZCR
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with bisphosphonate BPH-698 | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, tripotassium (1R)-4-(4'-butylbiphenyl-4-yl)-1-phosphonatobutane-1-sulfonate | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H, Oldfield, E. | | Deposit date: | 2007-11-11 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence.
Science, 319, 2008
|
|
