8IZG
 
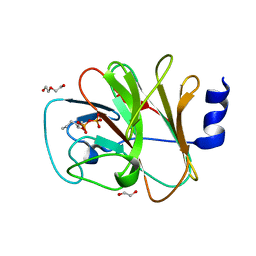 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with 5-HMBPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
1AVV
 
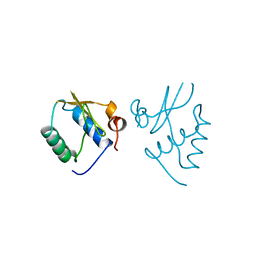 | | HIV-1 NEF PROTEIN, UNLIGANDED CORE DOMAIN | | Descriptor: | NEGATIVE FACTOR | | Authors: | Arold, S, Franken, P, Dumas, C. | | Deposit date: | 1997-09-21 | | Release date: | 1998-03-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling.
Structure, 5, 1997
|
|
1AVZ
 
 | |
5XEP
 
 | | Crystal structure of BRP39, a chitinase-like protein, at 2.6 Angstorm resolution | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-3-like protein 1 | | Authors: | Mohanty, A.K, Fisher, A.J, Choudhary, S, Kaushik, J.K. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of BRP39, a signalling glycoprotein expressed during mammary gland apoptosis.
To be published
|
|
6IJI
 
 | | Crystal structure of PDE10 in complex with inhibitor 2b | | Descriptor: | 2-{2-[5-methyl-1-(pyridin-4-yl)-1H-benzimidazol-2-yl]ethyl}-1H-benzo[de]isoquinoline-1,3(2H)-dione, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.Y, Yu, Y.F, Zhang, C, Wu, D, Wu, Y, Luo, H.B. | | Deposit date: | 2018-10-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Validation of Phosphodiesterase-10 as a Novel Target for Pulmonary Arterial Hypertension via Highly Selective and Subnanomolar Inhibitors.
J. Med. Chem., 62, 2019
|
|
6IJH
 
 | | Crystal structure of PDE10 in complex with inhibitor AF-399/14387019 | | Descriptor: | 2-[2-(4-phenyl-5-sulfanylidene-4,5-dihydro-1H-1,2,4-triazol-3-yl)ethyl]-1H-benzo[de]isoquinoline-1,3(2H)-dione, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.Y, Yu, Y.F, Zhang, C, Wu, D, Wu, Y, Luo, H.B. | | Deposit date: | 2018-10-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Validation of Phosphodiesterase-10 as a Novel Target for Pulmonary Arterial Hypertension via Highly Selective and Subnanomolar Inhibitors.
J. Med. Chem., 62, 2019
|
|
3IA3
 
 | |
7WZW
 
 | | Cryo-EM structure of MEC1-DDC2-MMS | | Descriptor: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | Authors: | Zhang, Q, Zhang, Q. | | Deposit date: | 2022-02-19 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of Mec1/ATR kinase endogenously stimulated by different genotoxins.
Cell Discov, 8, 2022
|
|
7WZR
 
 | | Cryo-EM structure of Mec1-HU | | Descriptor: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | Authors: | Zhang, Q, Zhang, Q. | | Deposit date: | 2022-02-19 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structures of Mec1/ATR kinase endogenously stimulated by different genotoxins.
Cell Discov, 8, 2022
|
|
2QZ2
 
 | | Crystal structure of a glycoside hydrolase family 11 xylanase from Aspergillus niger in complex with xylopentaose | | Descriptor: | Endo-1,4-beta-xylanase I, SODIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Vandermarliere, E, Rombouts, S, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
2Z79
 
 | | High resolution crystal structure of a glycoside hydrolase family 11 xylanase of Bacillus subtilis | | Descriptor: | Endo-1,4-beta-xylanase A, GLYCEROL | | Authors: | Vandermarliere, E, Bourgois, T.M, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
6YCQ
 
 | | Crystal structure of the DNA binding domain of Arabidopsis thaliana Auxin Response Factor 1 (AtARF1) in complex with High Affinity DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 21-7A, 21-7B, ... | | Authors: | Crespo, I, Weijers, D, Boer, D.R. | | Deposit date: | 2020-03-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2F6I
 
 | | Crystal structure of the ClpP protease catalytic domain from Plasmodium falciparum | | Descriptor: | ATP-dependent CLP protease, putative | | Authors: | Mulichak, A, Loppnau, P, Bray, J, Amani, M, Vedadi, M, Wasney, G, Finerty, P, Sundstrom, M, Weigelt, J, Edwards, A, Arrowsmith, C, Hui, R, Plotnikova, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Clp chaperones and proteases of the human malaria parasite Plasmodium falciparum.
J.Mol.Biol., 404, 2010
|
|
4QQZ
 
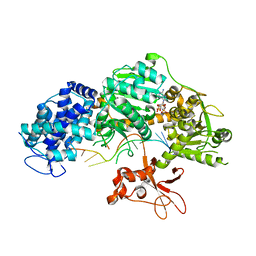 | | Crystal structure of T. fusca Cas3-AMPPNP | | Descriptor: | CRISPR-associated helicase, Cas3 family, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Ke, A, Huo, Y, Nam, K.H. | | Deposit date: | 2014-06-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4D8D
 
 | |
4QQY
 
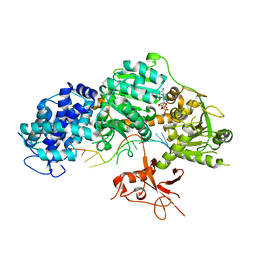 | | Crystal structure of T. fusca Cas3-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CRISPR-associated helicase, Cas3 family, ... | | Authors: | Ke, A, Huo, Y, Nam, K.H. | | Deposit date: | 2014-06-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4QQX
 
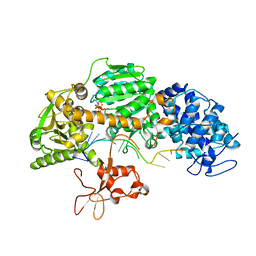 | | Crystal structure of T. fusca Cas3-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR-associated helicase, Cas3 family, ... | | Authors: | Ke, A, Huo, Y, Nam, K.H. | | Deposit date: | 2014-06-30 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4QQW
 
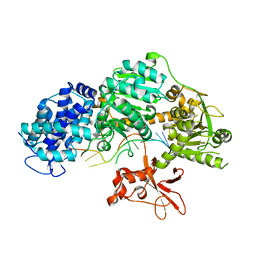 | | Crystal structure of T. fusca Cas3 | | Descriptor: | CRISPR-associated helicase, Cas3 family, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Ke, A, Huo, Y, Nam, K.H. | | Deposit date: | 2014-06-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.664 Å) | | Cite: | Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6GFT
 
 | |
1QA5
 
 | |
3E4A
 
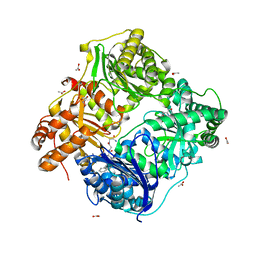 | | Human IDE-inhibitor complex at 2.6 angstrom resolution | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETIC ACID, HYDROXAMATE PEPTIDE II1, ... | | Authors: | Malito, E, Leissring, M.A, Choi, S, Cuny, G.D, Tang, W.J. | | Deposit date: | 2008-08-11 | | Release date: | 2009-05-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Designed inhibitors of insulin-degrading enzyme regulate the catabolism and activity of insulin.
Plos One, 5, 2010
|
|
1QA4
 
 | |
3PGG
 
 | | Crystal structure of cryptosporidium parvum u6 snrna-associated sm-like protein lsm5 | | Descriptor: | U6 snRNA-associated Sm-like protein LSm5. SM domain | | Authors: | Dong, A, Gao, M, Zhao, Y, Lew, J, Wasney, G.A, Kozieradzki, I, Vedadi, M, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-01 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Genome-Scale Protein Expression and Structural Biology of Plasmodium Falciparum and Related Apicomplexan Organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
4ORZ
 
 | |
5U08
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, ACETATE ION, CALCIUM ION, ... | | Authors: | Xu, Z, Skarina, T, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-23 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
