8YN8
 
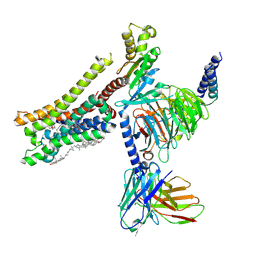 | | Cryo-EM structure of histamine H3 receptor in complex with proxyfan and miniGo | | Descriptor: | 5-(3-phenylmethoxypropyl)-1H-imidazole, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN4
 
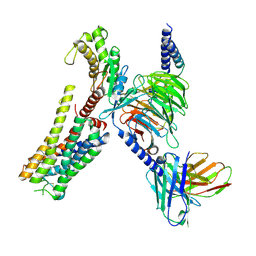 | | Cryo-EM structure of histamine H2 receptor in complex with histamine and miniGq | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Engineered guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN7
 
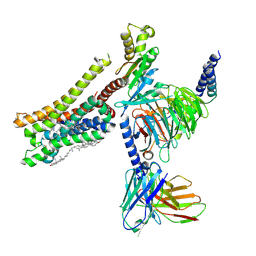 | | Cryo-EM structure of histamine H3 receptor in complex with immethridine and miniGo | | Descriptor: | 4-(1H-imidazol-4-ylmethyl)pyridine, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN6
 
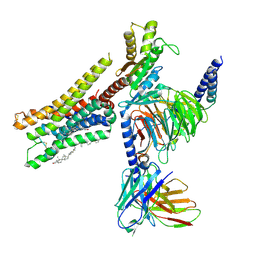 | | Cryo-EM structure of histamine H3 receptor in complex with imetit and Gi | | Descriptor: | 2-(1~{H}-imidazol-5-yl)ethyl carbamimidothioate, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN2
 
 | | Cryo-EM structure of histamine H1 receptor in complex with histamine and miniGq | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Engineered guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN9
 
 | | Cryo-EM structure of histamine H4 receptor in complex with histamine and Gi | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN5
 
 | | Cryo-EM structure of histamine H3 receptor in complex with histamine and Gi | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN3
 
 | | Cryo-EM structure of histamine H2 receptor in complex with histamine and miniGs | | Descriptor: | CHOLESTEROL, Engineered guanine nucleotide,binding protein G(s) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YNA
 
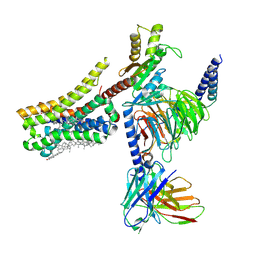 | | Cryo-EM structure of histamine H4 receptor in complex with immepip and Gi | | Descriptor: | 4-(1H-imidazol-5-ylmethyl)piperidine, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8I7X
 
 | | Crystal structure of human ClpP in complex with ZG36 | | Descriptor: | (6S,9aS)-N-[(4-bromophenyl)methyl]-6-[(2S)-butan-2-yl]-8-[(4-methoxynaphthalen-1-yl)methyl]-4,7-bis(oxidanylidene)-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Wang, P.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2023-02-02 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Assessment of the structure-activity relationship and antileukemic activity of diacylpyramide compounds as human ClpP agonists.
Eur.J.Med.Chem., 258, 2023
|
|
6KE3
 
 | |
6KDF
 
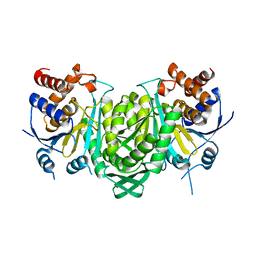 | | Crystal structure of the alpha beta heterodimer of human IDH3 in APO form. | | Descriptor: | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, Isocitrate dehydrogenase [NAD] subunit beta | | Authors: | Sun, P, Ding, J. | | Deposit date: | 2019-07-02 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Molecular basis for the function of the alpha beta heterodimer of human NAD-dependent isocitrate dehydrogenase.
J.Biol.Chem., 294, 2019
|
|
6KDE
 
 | |
6KDY
 
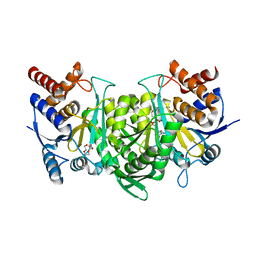 | |
5KIW
 
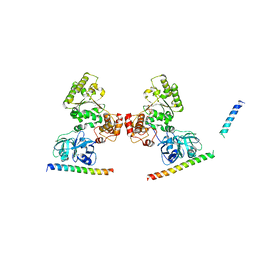 | | p97 ND1-L198W in complex with VIMP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Selenoprotein S, ... | | Authors: | Tang, W.K, Xia, D. | | Deposit date: | 2016-06-17 | | Release date: | 2018-03-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structural basis for nucleotide-modulated p97 association with the ER membrane.
Cell Discov, 3, 2017
|
|
5KIU
 
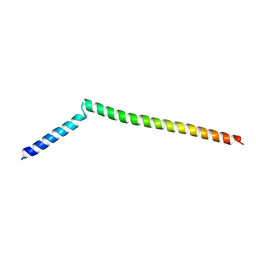 | | VCP-interacting membrane protein (VIMP) | | Descriptor: | Selenoprotein S | | Authors: | Tang, W.K, Xia, D. | | Deposit date: | 2016-06-17 | | Release date: | 2017-12-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for nucleotide-modulated p97 association with the ER membrane.
Cell Discov, 3, 2017
|
|
5KIY
 
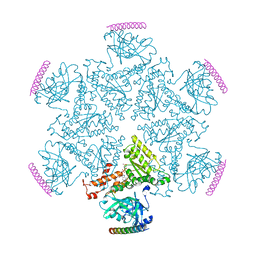 | | p97 ND1-A232E in complex with VIMP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Selenoprotein S, ... | | Authors: | Tang, W.K, Xia, D. | | Deposit date: | 2016-06-17 | | Release date: | 2017-12-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for nucleotide-modulated p97 association with the ER membrane.
Cell Discov, 3, 2017
|
|
8YZR
 
 | |
8Z07
 
 | |
8Z06
 
 | |
7XRD
 
 | |
8IGN
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with RAY1216 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S},3~{R})-4-(cyclopentylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8IGO
 
 | | Crystal structure of apo SARS-CoV-2 main protease | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8YZW
 
 | |
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
