7O13
 
 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7OSH
 
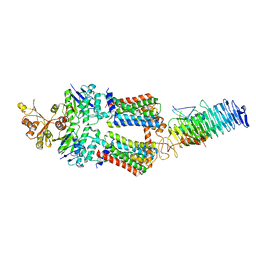 | | ABC Transporter complex NosDFYL, R-domain 2 | | Descriptor: | COPPER (II) ION, Copper-binding lipoprotein NosL, MAGNESIUM ION, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Du, J, Einsle, O. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7OSJ
 
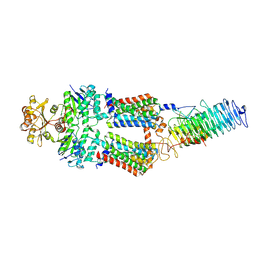 | | ABC Transporter complex NosDFYL, membrane anchor | | Descriptor: | COPPER (II) ION, Copper-binding lipoprotein NosL, MAGNESIUM ION, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Du, J, Einsle, O. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7OSI
 
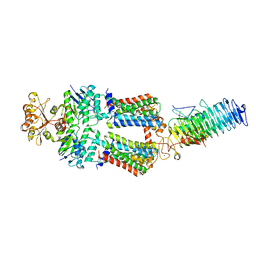 | | ABC Transporter complex NosDFYL, R-domain 3 | | Descriptor: | COPPER (II) ION, Copper-binding lipoprotein NosL, MAGNESIUM ION, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Du, J, Einsle, O. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7OSF
 
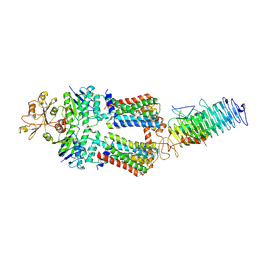 | | ABC Transporter complex NosDFYL, R-domain 1 | | Descriptor: | COPPER (II) ION, Copper-binding lipoprotein NosL, MAGNESIUM ION, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Du, J, Einsle, O. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7OSG
 
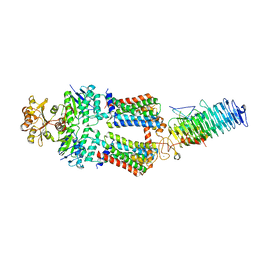 | | ABC Transporter complex NosDFYL, consensus refinement | | Descriptor: | COPPER (II) ION, Copper-binding lipoprotein NosL, MAGNESIUM ION, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Du, J, Einsle, O. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
5Y9H
 
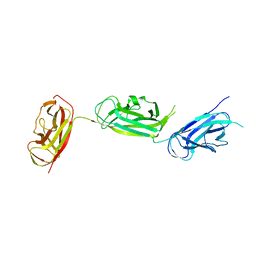 | | Crystal structure of SafDAA-dsc complex | | Descriptor: | SafD,Putative outer membrane protein,Putative outer membrane protein,Putative outer membrane protein | | Authors: | Zeng, L.H, Zhang, L, Wang, P.R, Meng, G. | | Deposit date: | 2017-08-25 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of host recognition and biofilm formation by Salmonella Saf pili
Elife, 6, 2017
|
|
5Y9G
 
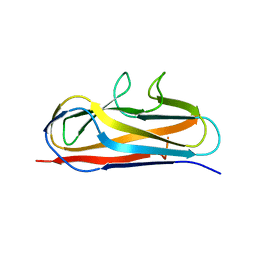 | | Crystal structure of Salmonella SafD adhesin | | Descriptor: | Pilin structural protein SafD,Pilus assembly protein | | Authors: | Zeng, L.H, Zhang, L, Wang, P.R, Meng, G. | | Deposit date: | 2017-08-25 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of host recognition and biofilm formation by Salmonella Saf pili
Elife, 6, 2017
|
|
6A67
 
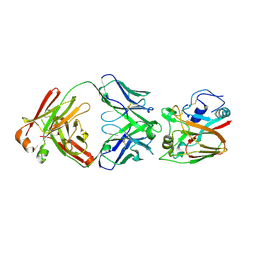 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody FLD21.140 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLD21.140 Heavy Chain, FLD21.140 Light Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Complementary recognition of the receptor-binding site of highly pathogenic H5N1 influenza viruses by two human neutralizing antibodies.
J. Biol. Chem., 293, 2018
|
|
9IWS
 
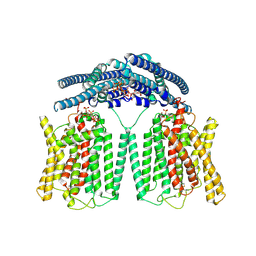 | | Cryo EM structure of human phosphate channel XPR1 in complex with IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-07-25 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 386, 2024
|
|
7RAV
 
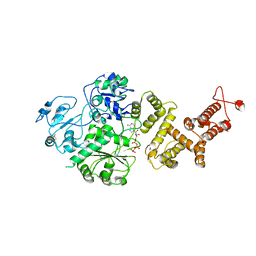 | | Cryo-EM structure of the unliganded form of NLR family apoptosis inhibitory protein 5 (NAIP5) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Baculoviral IAP repeat-containing protein 1e | | Authors: | Paidimuddala, B, Cao, J, Xie, Q, Wu, H, Zhang, L. | | Deposit date: | 2021-07-02 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of NAIP-NLRC4 inflammasome activation revealed by cryo-EM structure of unliganded NAIP5.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8VAU
 
 | | Nicotinamide Riboside and CD38: Covalent Inhibition and Live-Cell Labeling | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, Nicotinamide riboside, alpha-D-ribofuranose | | Authors: | Kao, G, Zhang, X.N, Nasertorabi, F, Katz, B.B, Li, Z, Dai, Z, Zhang, Z, Zhang, L, Louie, S.G, Cherezov, V, Zhang, Y. | | Deposit date: | 2023-12-11 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nicotinamide Riboside and CD38: Covalent Inhibition and Live-Cell Labeling.
Jacs Au, 4, 2024
|
|
8W72
 
 | |
8W6Z
 
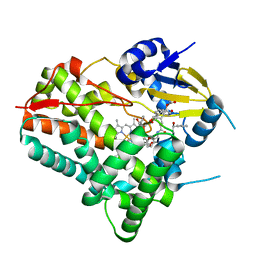 | | Substrate-bound crystal structure of a P450 enzyme DmlH that catalyze intramolecular phenol coupling in the biosynthesis of cihanmycins | | Descriptor: | (E)-3-[2-[(2R,3S)-3-[(1R)-1-aminocarbonyloxypropyl]oxiran-2-yl]phenyl]prop-2-enoic acid, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fang, C, Zhang, L, Zhu, Y, Zhang, C. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Biosynthesis of Cihanmycins Reveal Cytochrome P450-Catalyzed Intramolecular C-O Phenol Coupling Reactions.
J.Am.Chem.Soc., 2024
|
|
8QPF
 
 | |
5DUP
 
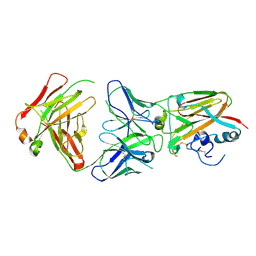 | | Influenza A virus H5 hemagglutinin globular head in complex with antibody AVFluIgG03 | | Descriptor: | AVFluIgG03 Heavy Chain, AVFluIgG03 Light Chain, Hemagglutinin | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUT
 
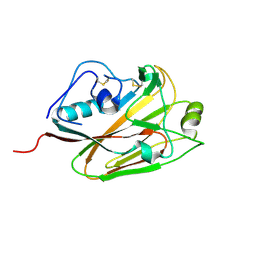 | | Influenza A virus H5 hemagglutinin globular head | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUM
 
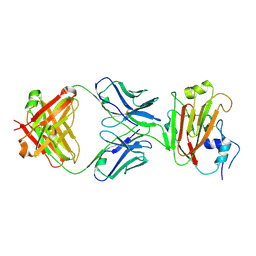 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody 65C6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 65C6 Heavy Chain, 65C6 Light Chain, ... | | Authors: | Sun, J, Zuo, T, Wang, G, Zhou, P, Zhang, L, Wang, X. | | Deposit date: | 2015-09-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUR
 
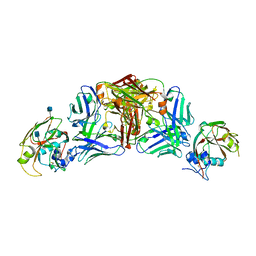 | | Influenza A virus H5 hemagglutinin globular head in complex with antibody 100F4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Antibody 100F4, Hemagglutinin, ... | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
7K26
 
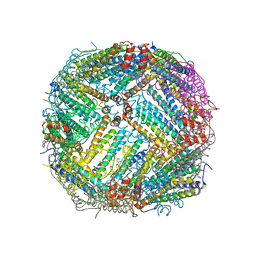 | |
7QBA
 
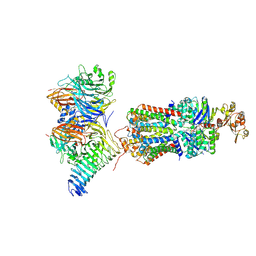 | | CryoEM structure of the ABC transporter NosDFY complexed with nitrous oxide reductase NosZ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zipfel, S, Mueller, C, Topitsch, A, Lutz, M, Zhang, L, Einsle, O. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
2MVT
 
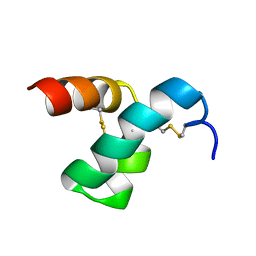 | | Solution structure of scoloptoxin SSD609 from Scolopendra mutilans | | Descriptor: | Scoloptoxin SSD609 | | Authors: | Wu, F, Sun, P, Wang, C, He, Y, Zhang, L, Tian, C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A distinct three-helix centipede toxin SSD609 inhibits Iks channels by interacting with the KCNE1 auxiliary subunit.
Sci Rep, 5, 2015
|
|
3WQL
 
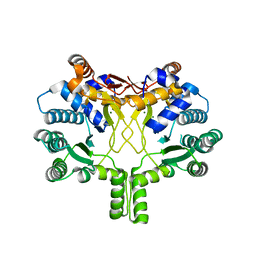 | | Crystal structure of Rv3378c with Mg2+ and PPi | | Descriptor: | Diterpene synthase, MAGNESIUM ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNF
 
 | | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
