8J49
 
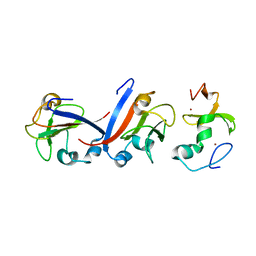 | | Crystal structure of OY phytoplasma SAP05 in complex with AtSPL5 | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8J48
 
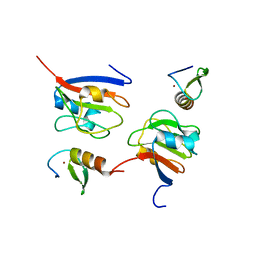 | | Crystal structure of OY phytoplasma SAP05 in complex with AtGATA18 | | Descriptor: | GATA transcription factor 18, Sequence-variable mosaic (SVM) signal sequence domain-containing protein, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
4YG3
 
 | |
5X5J
 
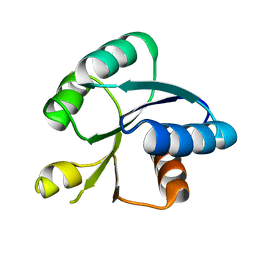 | | Crystal structure of response regulator AdeR receiver domain | | Descriptor: | AdeR | | Authors: | Wen, Y. | | Deposit date: | 2017-02-16 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region.
Nucleic Acids Res., 45, 2017
|
|
5XJP
 
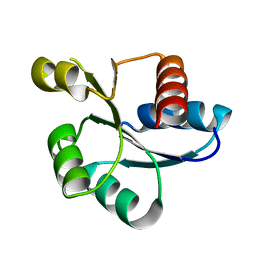 | | Crystal structure of response regulator AdeR receiver domain with Mg | | Descriptor: | AdeR, MAGNESIUM ION | | Authors: | Wen, Y. | | Deposit date: | 2017-05-03 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region.
Nucleic Acids Res., 45, 2017
|
|
5YGF
 
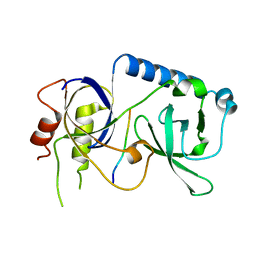 | |
5YGD
 
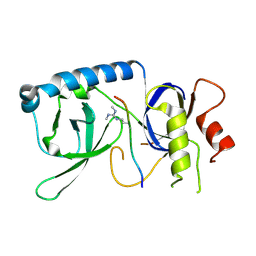 | |
5YGB
 
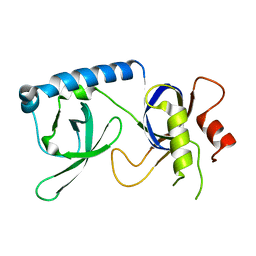 | |
5YGC
 
 | |
6IEW
 
 | |
8HI1
 
 | | Streptococcus thermophilus Cas1-Cas2- prespacer ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, DNA (26-MER), DNA (31-MER), ... | | Authors: | Chen, Q, Luo, Y. | | Deposit date: | 2022-11-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | DnaQ mediates directional spacer acquisition in the CRISPR-Cas system by a time-dependent mechanism.
Innovation (N Y), 4, 2023
|
|
6IHJ
 
 | |
3SR2
 
 | | Crystal Structure of Human XLF-XRCC4 Complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Hammel, M, Classen, S, Tainer, J.A. | | Deposit date: | 2011-07-06 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.9708 Å) | | Cite: | XRCC4 Protein Interactions with XRCC4-like Factor (XLF) Create an Extended Grooved Scaffold for DNA Ligation and Double Strand Break Repair.
J.Biol.Chem., 286, 2011
|
|
3U2Z
 
 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-(3-aminobenzyl)-4-methyl-2-methylsulfinyl-4,6-dihydro-5H-thieno[2',3':4,5]pyrrolo[2,3-d]pyridazin-5-one, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Nat.Chem.Biol., 8, 2012
|
|
7XIY
 
 | | SARS-CoV-2 Omicron BA.3 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7X6A
 
 | |
7XNQ
 
 | | SARS-CoV-2 Omicron BA.4 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIW
 
 | | SARS-CoV-2 Omicron BA.2 variant spike (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XNR
 
 | | SARS-CoV-2 Omicron BA.2.13 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XNS
 
 | | SARS-CoV-2 Omicron BA.4 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIZ
 
 | | SARS-CoV-2 Omicron BA.3 variant spike (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIX
 
 | | SARS-CoV-2 Omicron BA.2 variant spike (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7JU8
 
 | | X-ray structure of MMP-13 in Complex with 4-(1,2,3-thiadiazol-4-yl)pyridine | | Descriptor: | 4-(1,2,3-thiadiazol-4-yl)pyridine, CALCIUM ION, Collagenase 3, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2020-08-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Indole Inhibitors of MMP-13 for Arthritic Disorders
Acs Omega, 6, 2021
|
|
8HCI
 
 | |
8HI8
 
 | |
