5DEN
 
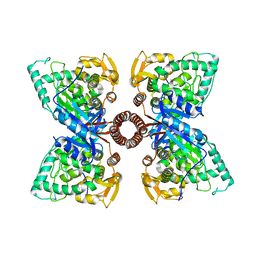 | |
3V46
 
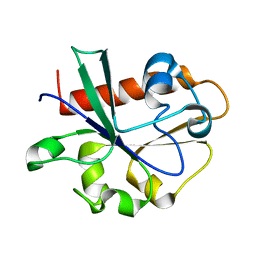 | | Crystal Structure of Yeast Cdc73 C-Terminal Domain | | Descriptor: | Cell division control protein 73 | | Authors: | Amrich, C.G, VanDemark, A.P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Cdc73 subunit of Paf1 complex contains C-terminal Ras-like domain that promotes association of Paf1 complex with chromatin.
J.Biol.Chem., 287, 2012
|
|
3F6P
 
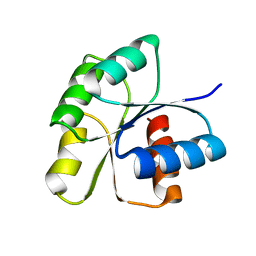 | | Crystal Structure of unphosphorelated receiver domain of YycF | | Descriptor: | Transcriptional regulatory protein yycF | | Authors: | Zhao, H, Tang, L. | | Deposit date: | 2008-11-06 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Preliminary crystallographic studies of the regulatory domain of response regulator YycF from an essential two-component signal transduction system.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
4QYZ
 
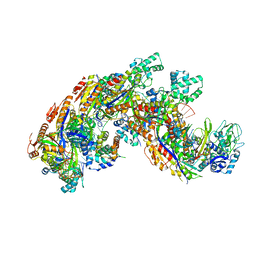 | | Crystal structure of a CRISPR RNA-guided surveillance complex, Cascade, bound to a ssDNA target | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Mulepati, S, Bailey, S. | | Deposit date: | 2014-07-26 | | Release date: | 2014-09-03 | | Last modified: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (3.0303 Å) | | Cite: | Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target.
Science, 345, 2014
|
|
1Z3Y
 
 | | Structure of Gun4-1 from Thermosynechococcus elongatus | | Descriptor: | putative cytidylyltransferase | | Authors: | Davison, P.A, Schubert, H.L, Reid, J.D, Iorg, C.D, Robinson, H, Hill, C.P, Hunter, C.N. | | Deposit date: | 2005-03-14 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Characterization of Gun4 Suggests a Mechanism for Its Role in Chlorophyll Biosynthesis(,).
Biochemistry, 44, 2005
|
|
1Z3X
 
 | | Structure of Gun4 from Thermosynechococcus elongatus | | Descriptor: | putative cytidylyltransferase | | Authors: | Davison, P.A, Schubert, H.L, Reid, J.D, Iorg, C.D, Robinson, H, Hill, C.P, Hunter, C.N. | | Deposit date: | 2005-03-14 | | Release date: | 2005-06-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Biochemical Characterization of Gun4 Suggests a Mechanism for Its Role in Chlorophyll Biosynthesis(,).
Biochemistry, 44, 2005
|
|
1RE1
 
 | |
4I6M
 
 | | Structure of Arp7-Arp9-Snf2(HSA)-RTT102 subcomplex of SWI/SNF chromatin remodeler. | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, PHOSPHATE ION, ... | | Authors: | Schubert, H.L, Cairns, B.R, Hill, C.P. | | Deposit date: | 2012-11-29 | | Release date: | 2013-02-13 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structure of an actin-related subcomplex of the SWI/SNF chromatin remodeler.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1RHQ
 
 | |
1RHJ
 
 | |
4IQ2
 
 | | P21 crystal form of FKBP12.6 | | Descriptor: | MALONIC ACID, Peptidyl-prolyl cis-trans isomerase FKBP1B | | Authors: | Chen, H, Mustafi, S.M, Li, H.M, LeMaster, D.M, Hernandez, G. | | Deposit date: | 2013-01-10 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and conformational flexibility of the unligated FK506-binding protein FKBP12.6.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IQC
 
 | | P3121 crystal form of FKBP12.6 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B | | Authors: | Chen, H, Mustafi, S.M, Li, H.M, LeMaster, D.M, Hernandez, G. | | Deposit date: | 2013-01-11 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure and conformational flexibility of the unligated FK506-binding protein FKBP12.6.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1RHM
 
 | |
1RHK
 
 | |
1RHR
 
 | |
1RHU
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF CASPASE-3 WITH A 5,6,7 TRICYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | (3S)-3-[({(2S)-5-[(N-ACETYL-L-ALPHA-ASPARTYL)AMINO]-4-OXO-1,2,4,5,6,7-HEXAHYDROAZEPINO[3,2,1-HI]INDOL-2-YL}CARBONYL)AMINO]-5-(BENZYLSULFANYL)-4-OXOPENTANOIC ACID, Caspase-3 | | Authors: | Becker, J.W, Rotonda, J, Soisson, S.M. | | Deposit date: | 2003-11-14 | | Release date: | 2004-05-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Reducing the Peptidyl Features of Caspase-3 Inhibitors: A Structural Analysis.
J.Med.Chem., 47, 2004
|
|
4MLA
 
 | | Structure of maize cytokinin oxidase/dehydrogenase 2 (ZmCKO2) | | Descriptor: | 1,2-ETHANEDIOL, Cytokinin oxidase 2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Morera, S, Kopecny, D, Briozzo, P, Koncitikova, R. | | Deposit date: | 2013-09-06 | | Release date: | 2015-03-11 | | Last modified: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
4L2W
 
 | |
4ML8
 
 | | Structure of maize cytokinin oxidase/dehydrogenase 2 (ZmCKO2) | | Descriptor: | Cytokinin oxidase 2, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Morera, S, Kopecny, D, Briozzo, P, Koncitikova, R. | | Deposit date: | 2013-09-06 | | Release date: | 2015-03-11 | | Last modified: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3LZS
 
 | | Crystal Structure of HIV-1 CRF01_AE Protease in Complex with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 protease | | Authors: | Schiffer, C.A, Bandaranayake, R.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
3LZU
 
 | | Crystal Structure of a Nelfinavir Resistant HIV-1 CRF01_AE Protease variant (N88S) in Complex with the Protease Inhibitor Darunavir. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 protease | | Authors: | Schiffer, C.A, Bandaranayake, R.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
3NPQ
 
 | |
3NPN
 
 | |
3LZV
 
 | | Structure of Nelfinavir-resistant HIV-1 protease (D30N/N88D) in complex with Darunavir. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 Protease, ... | | Authors: | Schiffer, C.A, Kolli, M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
4EUO
 
 | | Structure of Atu4243-GABA sensor | | Descriptor: | ABC transporter, substrate binding protein (Polyamine), GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Morera, S, Planamente, S. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural basis for selective GABA binding in bacterial pathogens.
Mol.Microbiol., 86, 2012
|
|
