4G63
 
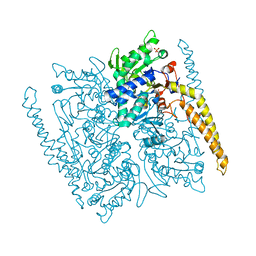 | | Crystal structure of cytosolic IMP-GMP specific 5'-nucleotidase (lpg0095) in complex with phosphate ions from Legionella pneumophila, Northeast Structural Genomics Consortium Target LgR1 | | Descriptor: | Cytosolic IMP-GMP specific 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Ho, C.K, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-07-18 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric regulation and substrate activation in cytosolic nucleotidase II from Legionella pneumophila.
Febs J., 281, 2014
|
|
1NXH
 
 | | X-RAY STRUCTURE: NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT87 | | Descriptor: | MTH396 protein | | Authors: | Khayat, R, Savchenko, A, Edwards, A, Arowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-10 | | Release date: | 2004-02-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-RAY STRUCTURE OF MTH396
To be Published
|
|
4FFD
 
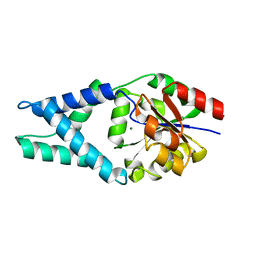 | | Crystal structure of engineered protein. northeast structural genomics consortium target or48 | | Descriptor: | MAGNESIUM ION, PF00702 DOMAIN PROTEIN | | Authors: | Seetharaman, J, Lew, S, Nivon, L, Baker, D, Bjelic, S, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of engineered protein. northeast structural genomics consortium target or48
To be Published
|
|
4FIB
 
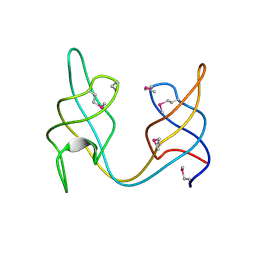 | | Crystal structure of the ydhK protein from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR518A. | | Descriptor: | Uncharacterized protein ydhK | | Authors: | Vorobiev, S, Abashidze, M, Seetharaman, J, Mao, M, Xiao, R, Maglaqui, M, Owens, L.A, Cunningham, K, Zhao, L, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-08 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structure of the ydhK protein from Bacillus subtilis.
To be Published
|
|
4FLE
 
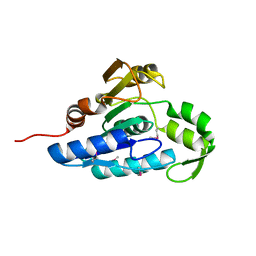 | | Crystal structure of the esterase YqiA (YE3661) from Yersinia enterocolitica, Northeast Structural Genomics Consortium Target YeR85 | | Descriptor: | esterase | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Shastry, R, Kohan, E, Maglaqui, M, Mao, L, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-14 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the esterase YqiA (YE3661) from Yersinia enterocolitica, Northeast Structural Genomics Consortium Target YeR85 (CASP Target)
To be Published
|
|
1NXU
 
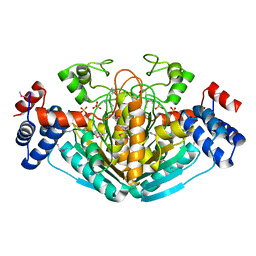 | | CRYSTAL STRUCTURE OF E. COLI HYPOTHETICAL OXIDOREDUCTASE YIAK NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER82. | | Descriptor: | Hypothetical oxidoreductase yiaK, SULFATE ION | | Authors: | Forouhar, F, Lee, I, Benach, J, Kulkarni, K, Xiao, R, Acton, T.B, Shastry, R, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel NAD-binding Protein Revealed by the Crystal Structure of 2,3-Diketo-L-gulonate Reductase (YiaK).
J.Biol.Chem., 279, 2004
|
|
1O77
 
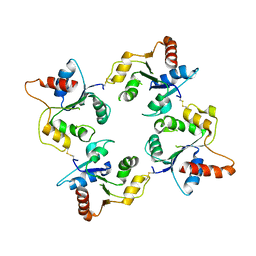 | | CRYSTAL STRUCTURE OF THE C713S MUTANT OF THE TIR DOMAIN OF HUMAN TLR2 | | Descriptor: | TOLL-LIKE RECEPTOR 2 | | Authors: | Tao, X, Xu, Y, Ye, Z, Beg, A.A, Tong, L. | | Deposit date: | 2002-10-24 | | Release date: | 2002-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | An Extensively Associated Dimer in the Structure of the C713S Mutant of the Tir Domain of Human Tlr2
Biochem.Biophys.Res.Commun., 299, 2002
|
|
1NXZ
 
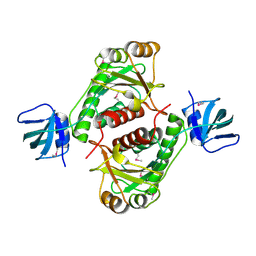 | | X-Ray Crystal Structure of Protein yggj_haein of Haemophilus influenzae. Northeast Structural Genomics Consortium Target IR73. | | Descriptor: | Hypothetical protein HI0303 | | Authors: | Forouhar, F, Shen, J, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional assignment based on structural analysis: crystal structure of the yggJ protein (HI0303) of Haemophilus influenzae reveals an RNA methyltransferase with a deep trefoil knot.
Proteins, 53, 2003
|
|
4F67
 
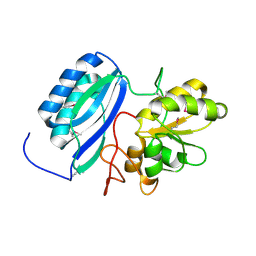 | | Three dimensional structure of the double mutant of UPF0176 protein lpg2838 from Legionella pneumophila at the resolution 1.8A, Northeast Structural Genomics Consortium (NESG) Target LgR82 | | Descriptor: | UPF0176 protein lpg2838 | | Authors: | Kuzin, A, Neely, H, Street, L, Odukwe, N, Seetharaman, J, Mao, M, Xiao, R, Kohan, E, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-14 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Three dimensional structure of the double mutant of UPF0176 protein lpg2838 from Legionella pneumophila at the resolution 1.8A, Northeast
Structural Genomics Consortium (NESG) Target LgR82
To be Published
|
|
1OD2
 
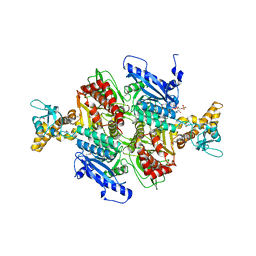 | | Acetyl-CoA Carboxylase Carboxyltransferase Domain | | Descriptor: | ACETYL COENZYME *A, ACETYL-COENZYME A CARBOXYLASE, ADENINE | | Authors: | Zhang, H, Yang, Z, Shen, Y, Tong, L. | | Deposit date: | 2003-02-12 | | Release date: | 2003-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase.
Science, 299, 2003
|
|
1OD4
 
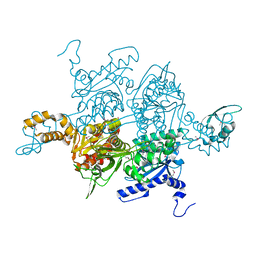 | | Acetyl-CoA Carboxylase Carboxyltransferase Domain | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE, ADENINE | | Authors: | Zhang, H, Yang, Z, Shen, Y, Tong, L. | | Deposit date: | 2003-02-12 | | Release date: | 2003-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase.
Science, 299, 2003
|
|
4DLH
 
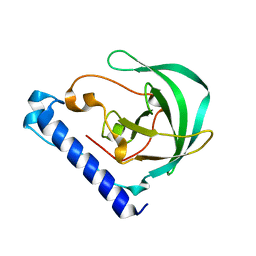 | | Crystal Structure of the protein Q9HRE7 from Halobacterium salinarium at the resolution 1.9A, Northeast Structural Genomics Consortium (NESG) Target HsR50 | | Descriptor: | uncharacterized protein | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target HsR50
To be Published
|
|
1OYZ
 
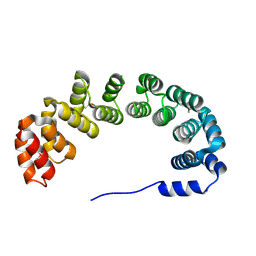 | | X-RAY STRUCTURE OF YIBA_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET31. | | Descriptor: | Hypothetical protein yibA | | Authors: | Kuzin, A, Semesi, A, Skarina, T, Savchenko, A, Arrowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-RAY STRUCTURE OF YIBA_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET31.
To be Published
|
|
1ON0
 
 | | Crystal Structure of Putative Acetyltransferase (YycN) from Bacillus subtilis, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR144 | | Descriptor: | CHLORIDE ION, SULFATE ION, YycN protein | | Authors: | Forouhar, F, Shen, J, Kuzin, A, Chiang, Y, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-26 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Putative Acetyltransferase (YycN) from Bacillus subtilis
To be Published
|
|
4PQ8
 
 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR465 | | Descriptor: | CHLORIDE ION, DESIGNED PROTEIN OR465 | | Authors: | Vorobiev, S, Parmeggiani, F, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Park, K, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-02-28 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Crystal Structure of Engineered Protein OR465.
To be Published
|
|
4PUT
 
 | |
4Q28
 
 | | Crystal Structure of the Plectin 1 and 2 Repeats of the Human Periplakin. Northeast Structural Genomics Consortium (NESG) Target HR9083A | | Descriptor: | Periplakin | | Authors: | Vorobiev, S, Lew, S, Seetharaman, J, Janjua, H, Xiao, R, O'Connell, P.T, Maglaqui, M, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal Structure of the Plectin 1 and 2 Repeats of Human Periplakin.
To be Published
|
|
4PWW
 
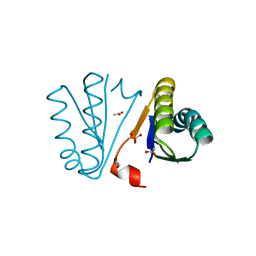 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR494. | | Descriptor: | ACETIC ACID, OR494, PHOSPHATE ION | | Authors: | Vorobiev, S, Lin, Y.-R, Seetharaman, J, Xiao, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Crystal Structure of Engineered Protein OR494.
To be Published
|
|
3U9T
 
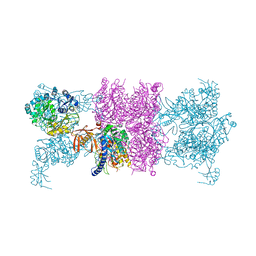 | |
3U13
 
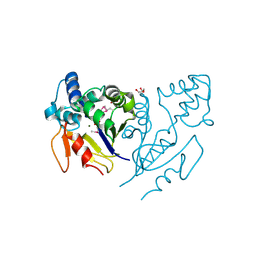 | | Crystal Structure of de Novo design of cystein esterase ECH13 at the resolution 1.6A, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3UAK
 
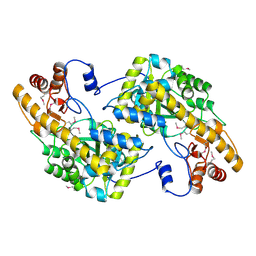 | | Crystal Structure of De Novo designed cysteine esterase ECH14, Northeast Structural Genomics Consortium Target OR54 | | Descriptor: | De Novo designed cysteine esterase ECH14 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-21 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.232 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3SYX
 
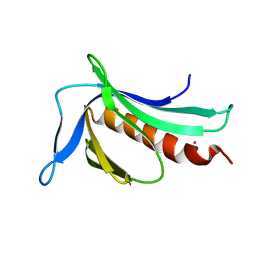 | | Crystal Structure of the WH1 domain from human sprouty-related, EVH1 domain-containing protein. Northeast Structural Genomics Consortium Target HR5538B. | | Descriptor: | Sprouty-related, EVH1 domain-containing protein 1, YTTRIUM (III) ION | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-18 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Crystal Structure of the WH1 domain from human sprouty-related, EVH1 domain-containing protein.
To be Published
|
|
4QIL
 
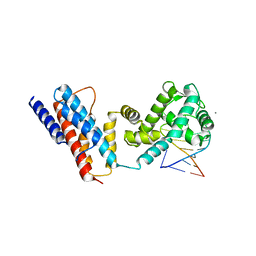 | |
3VB8
 
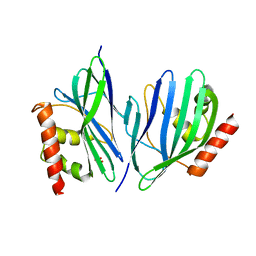 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR43 | | Descriptor: | Engineered protein, SULFATE ION | | Authors: | Seetharaman, J, Su, M, Procko, E, Baker, D, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-31 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Computational design of a protein-based enzyme inhibitor.
J.Mol.Biol., 425, 2013
|
|
4RHP
 
 | | Crystal structure of human COQ9 in complex with a phospholipid, Northeast Structural Genomics Consortium Target HR5043 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Ubiquinone biosynthesis protein COQ9, mitochondrial | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Wang, H, Lee, D, Kogan, S, Maglaqui, M, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
