1R3G
 
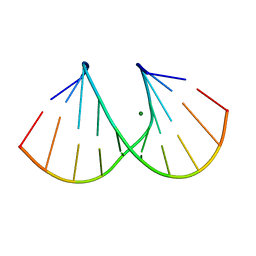 | | 1.16A X-ray structure of the synthetic DNA fragment with the incorporated 2'-O-[(2-Guanidinium)ethyl]-5-methyluridine residues | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(GMU)P*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Prakash, T.P, Puschl, A, Lesnik, E, Tereshko, V, Egli, M, Manoharan, M. | | Deposit date: | 2003-10-01 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 2'-O-[2-(Guanidinium)ethyl]-Modified Oligonucleotides: Stabilizing Effect on Duplex and Triplex Structures.
Org.Lett., 6, 2004
|
|
6JZI
 
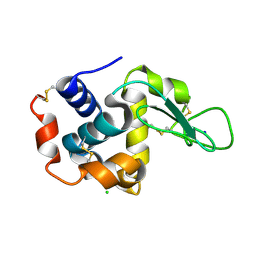 | | Structure of hen egg-white lysozyme obtained from SFX experiments under atmospheric pressure | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Nango, E, Sugahara, M, Nakane, T, Tanaka, T, Iwata, S. | | Deposit date: | 2019-05-02 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure.
J.Appl.Crystallogr., 52, 2019
|
|
1ZRN
 
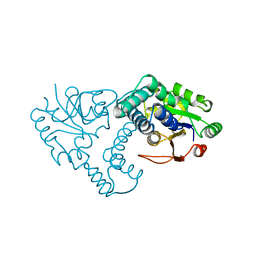 | | INTERMEDIATE STRUCTURE OF L-2-HALOACID DEHALOGENASE WITH MONOCHLOROACETATE | | Descriptor: | ACETIC ACID, L-2-HALOACID DEHALOGENASE | | Authors: | Li, Y.-F, Hata, Y, Fujii, T, Hisano, T, Nishihara, M, Kurihara, T, Esaki, N. | | Deposit date: | 1998-03-03 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of reaction intermediates of L-2-haloacid dehalogenase and implications for the reaction mechanism.
J.Biol.Chem., 273, 1998
|
|
1ZRM
 
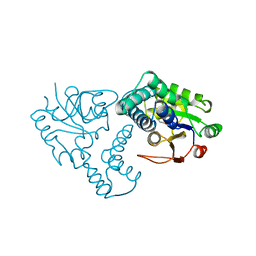 | | CRYSTAL STRUCTURE OF THE REACTION INTERMEDIATE OF L-2-HALOACID DEHALOGENASE WITH 2-CHLORO-N-BUTYRATE | | Descriptor: | L-2-HALOACID DEHALOGENASE, butanoic acid | | Authors: | Li, Y.-F, Hata, Y, Fujii, T, Hisano, T, Nishihara, M, Kurihara, T, Esaki, N. | | Deposit date: | 1998-03-03 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of reaction intermediates of L-2-haloacid dehalogenase and implications for the reaction mechanism.
J.Biol.Chem., 273, 1998
|
|
1YWN
 
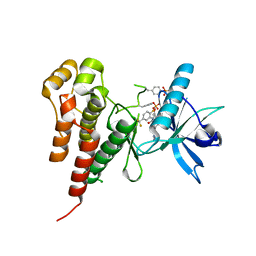 | | Vegfr2 in complex with a novel 4-amino-furo[2,3-d]pyrimidine | | Descriptor: | N-{4-[4-AMINO-6-(4-METHOXYPHENYL)FURO[2,3-D]PYRIMIDIN-5-YL]PHENYL}-N'-[2-FLUORO-5-(TRIFLUOROMETHYL)PHENYL]UREA, Vascular endothelial growth factor receptor 2 | | Authors: | Miyazaki, Y, Matsunaga, S, Tang, J, Maeda, Y, Nakano, M, Philippe, R.J, Shibahara, M, Liu, W, Sato, H, Wang, L, Nolte, R.T. | | Deposit date: | 2005-02-18 | | Release date: | 2005-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Novel 4-amino-furo[2,3-d]pyrimidines as Tie-2 and VEGFR2 dual inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2AXB
 
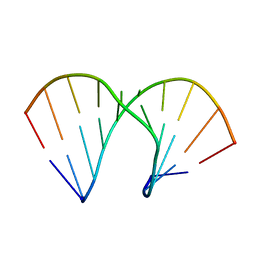 | | Crystal Structure Analysis Of A 2-O-[2-(methoxy)ethyl]-2-thiothymidine Modified Oligodeoxynucleotide Duplex | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(S2M)P*AP*CP*GP*C)-3') | | Authors: | Diop-Frimpong, B, Prakash, T.P, Rajeev, K.G, Manoharan, M, Egli, M. | | Deposit date: | 2005-09-04 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Stabilizing contributions of sulfur-modified nucleotides: crystal structure of a DNA duplex with 2'-O-[2-(methoxy)ethyl]-2-thiothymidines.
Nucleic Acids Res., 33, 2005
|
|
1U19
 
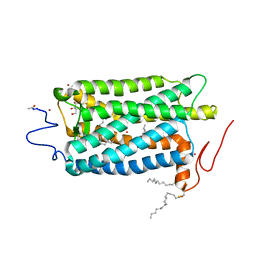 | | Crystal Structure of Bovine Rhodopsin at 2.2 Angstroms Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Okada, T, Sugihara, M, Bondar, A.N, Elstner, M, Entel, P, Buss, V. | | Deposit date: | 2004-07-15 | | Release date: | 2004-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure
J.Mol.Biol., 342, 2004
|
|
1ULR
 
 | | Crystal structure of tt0497 from Thermus thermophilus HB8 | | Descriptor: | putative acylphosphatase | | Authors: | Ago, H, Hamada, K, Sugahara, M, Kuroishi, C, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of tt0497 from Thermus thermophilus HB8
To be Published
|
|
1UJ4
 
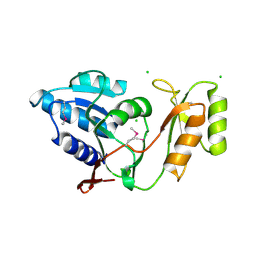 | | Crystal structure of Thermus thermophilus ribose-5-phosphate isomerase | | Descriptor: | CHLORIDE ION, ribose 5-phosphate isomerase | | Authors: | Hamada, K, Ago, H, Sugahara, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxyanion hole-stabilized stereospecific isomerization in ribose-5-phosphate isomerase (Rpi)
J.Biol.Chem., 278, 2003
|
|
1UJ6
 
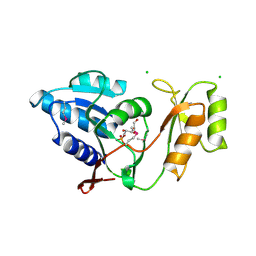 | | Crystal structure of Thermus thermophilus ribose-5-phosphate isomerase complexed with arabinose-5-phosphate | | Descriptor: | ARABINOSE-5-PHOSPHATE, CHLORIDE ION, ribose 5-phosphate isomerase | | Authors: | Hamada, K, Ago, H, Sugahara, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Oxyanion hole-stabilized stereospecific isomerization in ribose-5-phosphate isomerase (Rpi)
J.Biol.Chem., 278, 2003
|
|
1UJ5
 
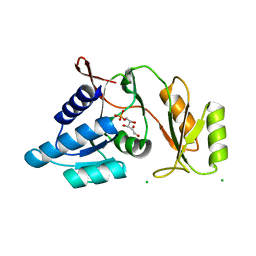 | | Crystal structure of Thermus thermophilus ribose-5-phosphate isomerase complexed with ribose-5-phosphate | | Descriptor: | CHLORIDE ION, RIBULOSE-5-PHOSPHATE, ribose 5-phosphate isomerase | | Authors: | Hamada, K, Ago, H, Sugahara, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxyanion hole-stabilized stereospecific isomerization in ribose-5-phosphate isomerase (Rpi)
J.Biol.Chem., 278, 2003
|
|
1ULS
 
 | | Crystal structure of tt0140 from Thermus thermophilus HB8 | | Descriptor: | putative 3-oxoacyl-acyl carrier protein reductase | | Authors: | Hisanaga, Y, Ago, H, Hamada, K, Sugahara, M, Nodake, Y, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of tt0140 from Thermus thermophilus HB8
To be Published
|
|
4QCD
 
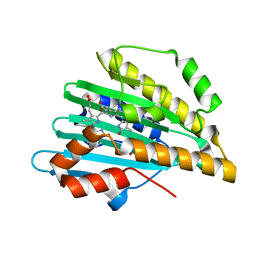 | | Neutron crystal structure of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin IXalpha at room temperature. | | Descriptor: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase, trideuteriooxidanium | | Authors: | Unno, M, Ishikawa-Suto, K, Ishihara, M, Hagiwara, Y, Sugishima, M, Wada, K, Fukuyama, K. | | Deposit date: | 2014-05-10 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (1.932 Å), X-RAY DIFFRACTION | | Cite: | Insights into the Proton Transfer Mechanism of a Bilin Reductase PcyA Following Neutron Crystallography.
J. Am. Chem. Soc., 137, 2015
|
|
1J2T
 
 | | Creatininase Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, ZINC ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1J2U
 
 | | Creatininase Zn | | Descriptor: | SULFATE ION, ZINC ION, creatinine amidohydrolase | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
3VU9
 
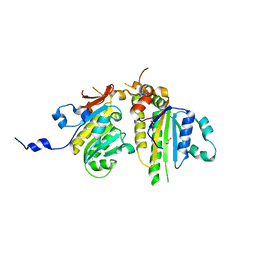 | | Crystal Structure of Psy3-Csm2 complex | | Descriptor: | 1,2-ETHANEDIOL, Chromosome segregation in meiosis protein 2, Platinum sensitivity protein 3 | | Authors: | Tawaramoto, M, Sasanuma, H, Hosaka, H, Lao, J.P, Sanda, E, Suzuki, M, Yamashita, E, Hunter, N, Shinohara, M, Nakagawa, A, Shinohara, A. | | Deposit date: | 2012-06-23 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A new protein complex promoting the assembly of Rad51 filaments
Nat Commun, 4, 2013
|
|
5AXD
 
 | |
5AXA
 
 | |
5AXC
 
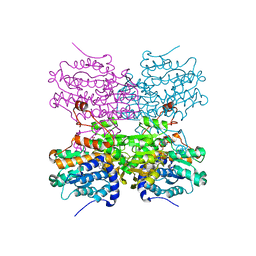 | | Crystal structure of mouse SAHH complexed with 3'-keto aristeromycin | | Descriptor: | (2S,3R,5R)-3-(6-amino-9H-purin-9-yl)-2-hydroxy-5-(hydroxymethyl)cyclopentanone, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kusakabe, Y, Ishihara, M, Tanaka, N. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of mouse SAHH complexed with 3'-keto aristeromycin
To Be Published
|
|
5AXB
 
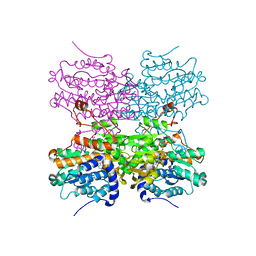 | | Crystal structure of mouse SAHH complexed with noraristeromycin | | Descriptor: | (1S,2R,3S,4R)-4-(6-aminopurin-9-yl)cyclopentane-1,2,3-triol, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kusakabe, Y, Ishihara, M, Tanaka, N. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of mouse SAHH complexed with noraristeromycin
To Be Published
|
|
1TYN
 
 | | ATOMIC STRUCTURE OF THE TRYPSIN-CYCLOTHEONAMIDE A COMPLEX: LESSONS FOR THE DESIGN OF SERINE PROTEASE INHIBITORS | | Descriptor: | BETA-TRYPSIN, CYCLOTHEONAMIDE A | | Authors: | Lee, A.Y, Hagihara, M, Karmacharya, R, Albers, M.W, Schreiber, S.L, Clardy, J. | | Deposit date: | 1994-09-19 | | Release date: | 1995-01-26 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic Structure of the Trypsin-Cyclotheonamide a Complex: Lessons for the Design of Serine Protease Inhibitors
J.Am.Chem.Soc., 115, 1993
|
|
1V7Z
 
 | | creatininase-product complex | | Descriptor: | MANGANESE (II) ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-12-26 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
5HS3
 
 | | Human thymidylate synthase complexed with dUMP and 3-amino-2-benzoyl-4-methylthieno[2,3-b]pyridin-6-ol | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 3-amino-2-benzoyl-4-methylthieno[2,3-b]pyridin-6-ol, Thymidylate synthase | | Authors: | Chen, D, Almqvist, H, Axelsson, H, Jafari, R, Mateus, A, Haraldsson, M, Larsson, A, Artursson, P, Molina, D.M, Lundback, T, Nordlund, P. | | Deposit date: | 2016-01-25 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | CETSA screening identifies known and novel thymidylate synthase inhibitors and slow intracellular activation of 5-fluorouracil
Nat Commun, 7, 2016
|
|
2NOO
 
 | | Crystal Structure of Mutant NikA | | Descriptor: | IODIDE ION, NICKEL (II) ION, Nickel-binding periplasmic protein | | Authors: | Addy, C, Ohara, M, Kawai, F, Kidera, A, Ikeguchi, M, Fuchigami, S, Osawa, M, Shimada, I, Park, S.Y, Tame, J.R.H, Heddle, J.G. | | Deposit date: | 2006-10-26 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nickel binding to NikA: an additional binding site reconciles spectroscopy, calorimetry and crystallography.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2DCT
 
 | | Crystal structure of the TT1209 from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, SODIUM ION, hypothetical protein TTHA0104 | | Authors: | Asada, Y, Sugahara, M, Shimizu, K, Yamamoto, H, Shimada, H, Nakamoto, T, Ono, N, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-12 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the TT1209 from Thermus thermophilus HB8
To be Published
|
|
