8FB6
 
 | | Crystal structure of Ky15.10 Fab in complex with circumsporozoite protein KQPA peptide | | Descriptor: | 1,2-ETHANEDIOL, Circumsporozoite protein KQPA peptide, Ky15.10 Antibody, ... | | Authors: | Prieto, K, Thai, E, Julien, J.P. | | Deposit date: | 2022-11-29 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
8FBA
 
 | |
8F9U
 
 | |
8FA8
 
 | |
8FAS
 
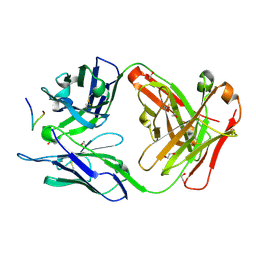 | | Crystal structure of Ky230 Fab in complex with circumsporozoite protein NANP5 peptide | | Descriptor: | 1,2-ETHANEDIOL, Circumsporozoite protein NANP5 peptide, Ky230 Antibody, ... | | Authors: | Kassardjian, A, Thai, E, Julien, J.P. | | Deposit date: | 2022-11-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
8FDC
 
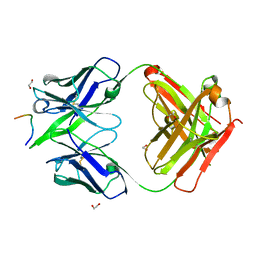 | |
6EG3
 
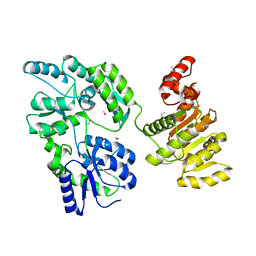 | | Crystal structure of human BRM in complex with compound 15 | | Descriptor: | 3-[(4-{[(2-chloropyridin-4-yl)carbamoyl]amino}pyridin-2-yl)ethynyl]benzoic acid, ETHANOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2 | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
6EG2
 
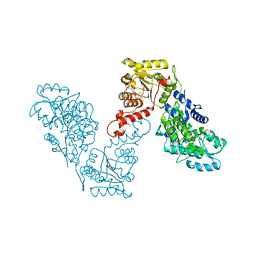 | | Crystal structure of human BRM in complex with compound 16 | | Descriptor: | ISOPROPYL ALCOHOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2, N-(5-amino-2-chloropyridin-4-yl)-N'-(4-bromo-3-{[3-(hydroxymethyl)phenyl]ethynyl}-1,2-thiazol-5-yl)urea | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
8C9J
 
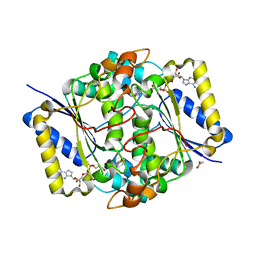 | | Crystal structure of human NQO1 by serial femtosecond crystallography | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Martin-Garcia, J.M, Grieco, A, Ruiz-Fresneda, M.A, Pacheco-Garcia, J.L, Pey, A, Botha, S, Ros, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modular droplet injector for sample conservation providing new structural insight for the conformational heterogeneity in the disease-associated NQO1 enzyme.
Lab Chip, 23, 2023
|
|
4QQS
 
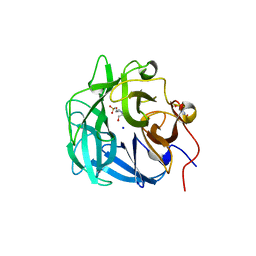 | | Crystal structure of a thermostable family-43 glycoside hydrolase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycoside hydrolase family 43, SODIUM ION | | Authors: | Hassan, N, Kori, L.D, Patel, B.K.C, Divne, C, Tan, T.C. | | Deposit date: | 2014-06-29 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution crystal structure of a polyextreme GH43 glycosidase from Halothermothrix orenii with alpha-L-arabinofuranosidase activity.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
6CDX
 
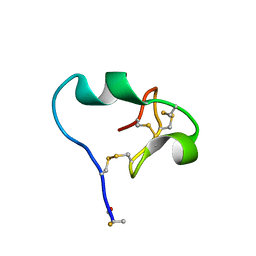 | | High-resolution crystal structure of fluoropropylated cystine knot, binding to alpha-5 beta-6 integrin | | Descriptor: | cystine knot (fluoropropylated) | | Authors: | Kimura, R, Nix, J, Bongura, C, Chakraborti, S, Gambhir, S, Filipp, F.V. | | Deposit date: | 2018-02-09 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Evaluation of integrin alpha v beta6cystine knot PET tracers to detect cancer and idiopathic pulmonary fibrosis.
Nat Commun, 10, 2019
|
|
8FG0
 
 | |
7DVK
 
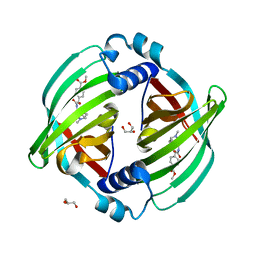 | | PyrI4 in complex with intermolecular Diels-Alder product | | Descriptor: | GLYCEROL, Spiro-conjugate synthase, methyl 4-[[(1S,6S)-6-(dimethylcarbamoyl)cyclohex-2-en-1-yl]carbamoyloxymethyl]benzoate | | Authors: | Kashyap, R, Addlagatta, A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exo-selective intermolecular Diels-Alder reaction by PyrI4 and AbnU on non-natural substrates.
Commun Chem, 2021
|
|
7DVI
 
 | |
7BBJ
 
 | |
3CVZ
 
 | |
7RCL
 
 | | Crystal Structure of ADP-bound Galactokinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Galactokinase, MAGNESIUM ION, ... | | Authors: | Whitby, F.G, Hall, M.D. | | Deposit date: | 2021-07-07 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Optimization of Small Molecule Human Galactokinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7RCM
 
 | | Crystal Structure of ADP-bound Galactokinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Galactokinase, ... | | Authors: | Whitby, F.G. | | Deposit date: | 2021-07-07 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Optimization of Small Molecule Human Galactokinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
6MDA
 
 | |
7S49
 
 | | Crystal Structure of Inhibitor-bound Galactokinase | | Descriptor: | (4R)-2-[(1,3-benzoxazol-2-yl)amino]-4-(4-chloro-1H-pyrazol-5-yl)-4,6,7,8-tetrahydroquinazolin-5(1H)-one, Galactokinase, PHOSPHATE ION, ... | | Authors: | Whitby, F.G. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization of Small Molecule Human Galactokinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7S4C
 
 | | Crystal Structure of Inhibitor-bound Galactokinase | | Descriptor: | 2-({(4R)-4-(2-chlorophenyl)-2-[(6-fluoro-1,3-benzoxazol-2-yl)amino]-6-methyl-1,4-dihydropyrimidine-5-carbonyl}amino)pyridine-4-carboxylic acid, Galactokinase, PHOSPHATE ION, ... | | Authors: | Whitby, F.G. | | Deposit date: | 2021-09-08 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization of Small Molecule Human Galactokinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
6MD9
 
 | |
1VKQ
 
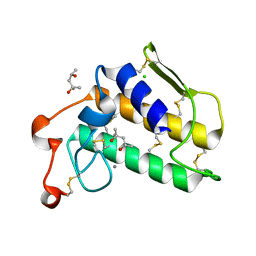 | | A re-determination of the structure of the triple mutant (K53,56,120M) of phospholipase A2 at 1.6A resolution using sulphur-SAS at 1.54A wavelength | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sekar, K, Velmurugan, D, Rajakannan, V, Yamane, T, Dauter, M, Dauter, Z. | | Deposit date: | 2004-06-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A redetermination of the structure of the triple mutant (K53,56,120M) of phospholipase A2 at 1.6 A resolution using sulfur-SAS at 1.54 A wavelength.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6MDD
 
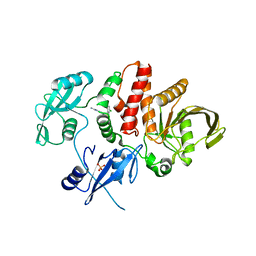 | |
6MDC
 
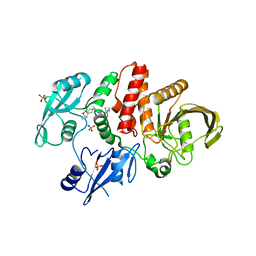 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor Pyrazolo-pyrimidinone 1 SHP389 | | Descriptor: | 6-[(3S,4S)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-[3-chloro-2-(cyclopropylamino)pyridin-4-yl]-5-methyl-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Fodor, M, Stams, T. | | Deposit date: | 2018-09-04 | | Release date: | 2019-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Optimization of Fused Bicyclic Allosteric SHP2 Inhibitors.
J. Med. Chem., 62, 2019
|
|
