7WAE
 
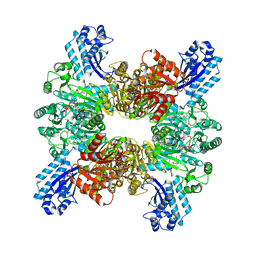 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS, 4x(beta-Asp-Arg), and aspartate | | Descriptor: | 4x(beta-Asp-Arg), ARGININE, ASPARTIC ACID, ... | | Authors: | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
1NI7
 
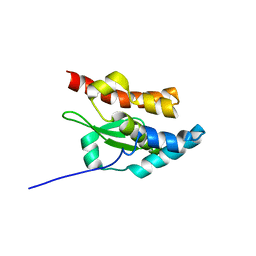 | | NORTHEAST STRUCTURAL GENOMIC CONSORTIUM TARGET ER75 | | Descriptor: | Hypothetical protein ygdK | | Authors: | Liu, G, Chiang, Y, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-21 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-quality homology models derived from NMR and X-ray structures of E. coli proteins YgdK and Suf E suggest that all members of the YgdK/Suf E protein family are enhancers of cysteine desulfurases.
Protein Sci., 14, 2005
|
|
7VU6
 
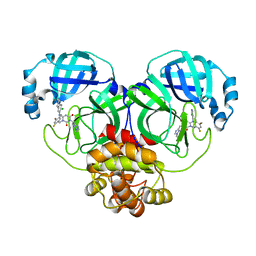 | | The crystal structure of SARS-CoV-2 3CL protease in complex with compound 3 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Yamamoto, S, Yamane, J, Tachibana, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19.
J.Med.Chem., 65, 2022
|
|
7VTH
 
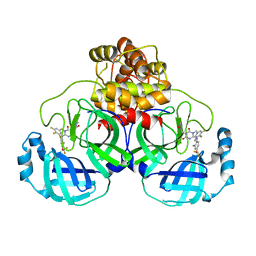 | | The crystal structure of SARS-CoV-2 3CL protease in complex with compound 1 | | Descriptor: | 2-[4-[[4-[bis(fluoranyl)methoxy]-2-methyl-phenyl]amino]-2,6-bis(oxidanylidene)-3-[[3,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazin-1-yl]-N-methyl-ethanamide, 3C-like proteinase | | Authors: | Yamamoto, S, Tachibana, Y. | | Deposit date: | 2021-10-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19.
J.Med.Chem., 65, 2022
|
|
1NY1
 
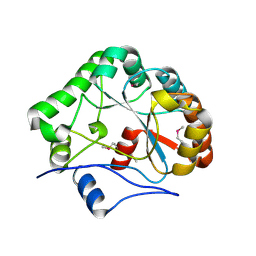 | | CRYSTAL STRUCTURE OF B. SUBTILIS POLYSACCHARIDE DEACETYLASE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR127. | | Descriptor: | Probable polysaccharide deacetylase pdaA | | Authors: | Forouhar, F, Edstrom, W, Khan, J, Ma, L, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Polysaccharide Deacetylase (PDAA_BACSU) from B. Subtilis (Pdaa_Bacsu) Northeast Structural Genomics Research Consortium (Nesg) Target Sr127
To be Published
|
|
3VQU
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 4-[(4-amino-5-cyano-6-ethoxypyridin-2- yl)amino]benzamide | | Descriptor: | 4-[(4-amino-5-cyano-6-ethoxypyridin-2-yl)amino]benzamide, Dual specificity protein kinase TTK, IODIDE ION | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Higashino, K, Okano, Y, Tadano, G, Tachibana, Y, Sato, Y, Inoue, M, Wada, T, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Tagashira, S, Kido, Y, Sakamoto, S, Yasuo, K, Maeda, M, Yamamoto, T, Higaki, M, Endoh, T, Ueda, K, Shiota, T, Murai, H, Nakamura, Y. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diaminopyridine-based potent and selective mps1 kinase inhibitors binding to an unusual flipped-Peptide conformation.
Acs Med.Chem.Lett., 3, 2012
|
|
6ABU
 
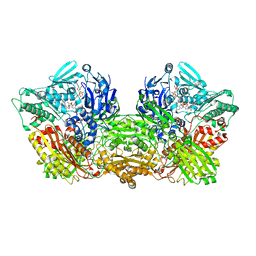 | | Rat Xanthine oxidoreductase, NAD bound form | | Descriptor: | BICARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Rat Xanthine oxidoreductase, NAD bound form
To Be Published
|
|
1L7Y
 
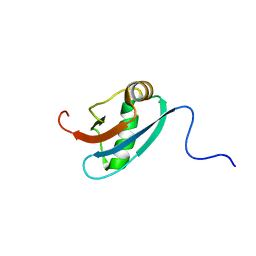 | | Solution NMR Structure of C. elegans Protein ZK652.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR41. | | Descriptor: | HYPOTHETICAL PROTEIN ZK652.3 | | Authors: | Cort, J.R, Chiang, Y, Zheng, D, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-03-18 | | Release date: | 2002-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of conserved eukaryotic protein ZK652.3 from C. elegans: a ubiquitin-like fold.
Proteins, 48, 2002
|
|
3VS1
 
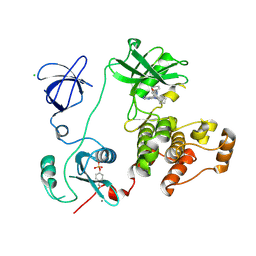 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-phenylurea | | Descriptor: | 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-phenylurea, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.464 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
5XLH
 
 | | Crystal structure of aerobically purified and aerobically crystallized for 12weeks D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
5XLG
 
 | | Crystal structure of anaerobically purified and aerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
7E5O
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with antibody NT-193 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-193 Heavy chain, NT-193 Light chain, ... | | Authors: | Kita, S, Onodera, T, Adachi, Y, Moriayma, S, Nomura, T, Tadokoro, T, Anraku, Y, Yumoto, K, Tian, C, Fukuhara, H, Suzuki, T, Tonouchi, K, Sasaki, J, Sun, L, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 antibody broadly neutralizes SARS-related coronaviruses and variants by coordinated recognition of a virus-vulnerable site.
Immunity, 54, 2021
|
|
3VS4
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 5-(4-phenoxyphenyl)-7-(tetrahydro-2H-pyran-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 5-(4-phenoxyphenyl)-7-(tetrahydro-2H-pyran-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.747 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
1NPD
 
 | | X-RAY STRUCTURE OF SHIKIMATE DEHYDROGENASE COMPLEXED WITH NAD+ FROM E.COLI (YDIB) NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER24 | | Descriptor: | HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Benach, J, Kuzin, A.P, Lee, I, Rost, B, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-17 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 2.3-A crystal structure of the shikimate 5-dehydrogenase orthologue YdiB from Escherichia coli suggests a novel catalytic environment for an NAD-dependent dehydrogenase
J.Biol.Chem., 278, 2003
|
|
5XYO
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122G mutant | | Descriptor: | CHLORIDE ION, Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, ... | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYP
 
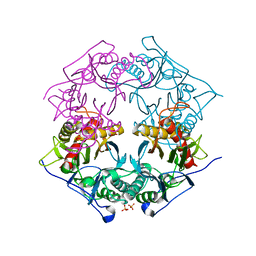 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122R mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYT
 
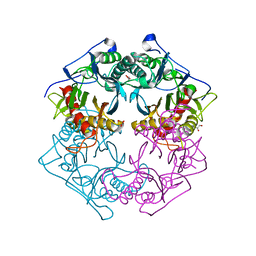 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., H130Y mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, SULFATE ION | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
3W1F
 
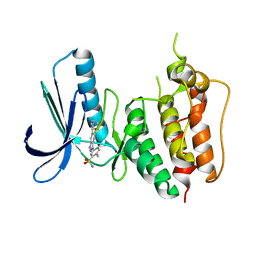 | | Crystal structure of Human MPS1 catalytic domain in complex with 5-(5-ethoxy-6-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-3-yl)-2-methylbenzenesulfonamide | | Descriptor: | 5-[5-ethoxy-6-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-3-yl]-2-methylbenzenesulfonamide, Dual specificity protein kinase TTK | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Tachibana, Y, Itoh, T, Yamamoto, T, Hashizume, H, Hato, Y, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Yasuo, K, Maeda, M, Higaki, M, Ueda, K, Yoshizawa, H, Baba, Y, Shiota, T, Murai, H, Nakamura, Y. | | Deposit date: | 2012-11-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Indazole-based potent and cell-active Mps1 kinase inhibitors: rational design from pan-kinase inhibitor anthrapyrazolone (SP600125)
J.Med.Chem., 56, 2013
|
|
3VS3
 
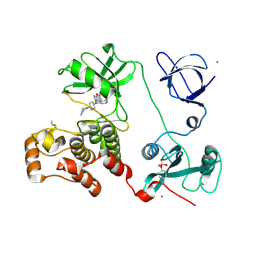 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomaebchi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS0
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor N-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]benzamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, N-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]benzamide, ... | | Authors: | Kuratani, M, Honda, K, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.934 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
4E9R
 
 | | Multicopper Oxidase CueO (data4) | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2012-03-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site.
J.Mol.Biol., 373, 2007
|
|
3VRY
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane | | Descriptor: | 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VRZ
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-benzylurea | | Descriptor: | 1-[4-(4-amino-7-cyclopentyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]-3-benzylurea, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS5
 
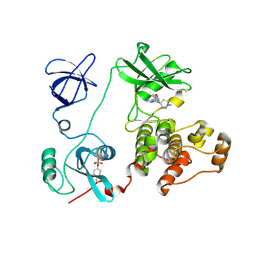 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-(1-methylpiperidin-4-yl)-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, Tyrosine-protein kinase HCK | | Authors: | Kuratani, M, Tomabechi, Y, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3VS6
 
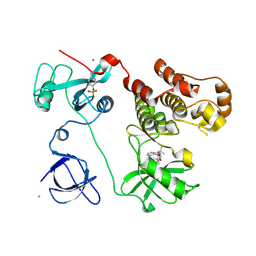 | | Crystal structure of HCK complexed with a pyrazolo-pyrimidine inhibitor tert-butyl {4-[4-amino-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl]-2-methoxyphenyl}carbamate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tyrosine-protein kinase HCK, ... | | Authors: | Kuratani, M, Honda, K, Tomabechi, Y, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.373 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
