3SWI
 
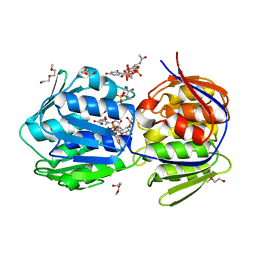 | | E. Cloacae MurA in complex with Enolpyruvyl-UDP-N-acetylgalactosamine and covalent adduct of PEP with CYS115 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{[(2R,3R,4R,5R,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}prop-2-enoic acid, MAGNESIUM ION, ... | | Authors: | Zhu, J.-Y, Yang, Y, Schonbrunn, E. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
3SWQ
 
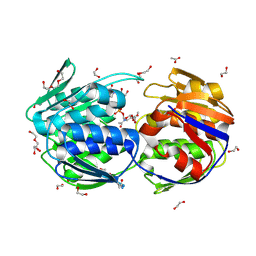 | | E. Cloacae MurA in complex with Enolpyruvyl-UNAG | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhu, J.-Y, Yang, Y, Schonbrunn, E. | | Deposit date: | 2011-07-14 | | Release date: | 2012-03-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
3SWD
 
 | | E. coli MurA in complex with UDP-N-acetylmuramic acid and covalent adduct of PEP with Cys115 | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
6V0Q
 
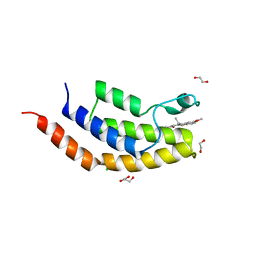 | | Crystal structure of the bromodomain of human BRD7 bound to TG003 | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 7, ... | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V0S
 
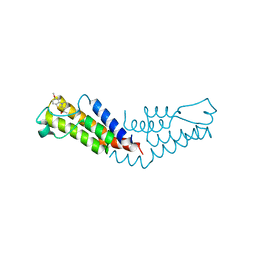 | |
6V0X
 
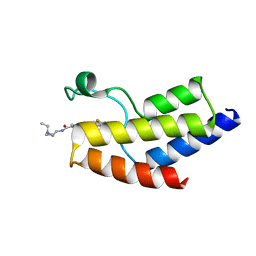 | | Crystal structure of the bromodomain of human BRD9 bound to sunitinib | | Descriptor: | Bromodomain-containing protein 9, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Karim, M.R, Chan, A, Zhu, J.Y, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1K
 
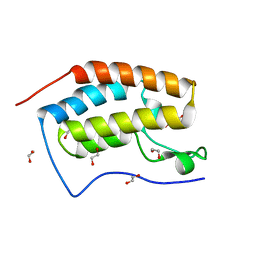 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to BI7273 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-[(dimethylamino)methyl]-3,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 4 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V16
 
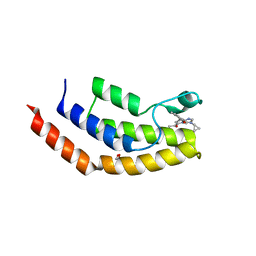 | | Crystal structure of the bromodomain of human BRD7 bound to TP472 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-acetylpyrrolo[1,2-a]pyrimidin-8-yl)-N-cyclopropyl-4-methylbenzamide, Bromodomain-containing protein 7, ... | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1H
 
 | | Crystal structure of the bromodomain of human BRD7 bound to bromosporine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 7, Bromosporine | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1L
 
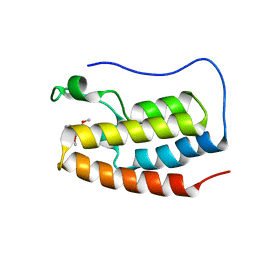 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to BI9564 | | Descriptor: | 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 4 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1U
 
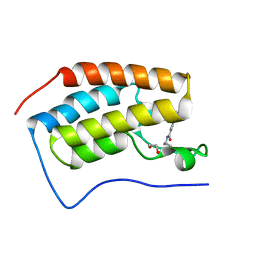 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to TP-472 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-acetylpyrrolo[1,2-a]pyrimidin-8-yl)-N-cyclopropyl-4-methylbenzamide, Bromodomain-containing protein 4 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-21 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V14
 
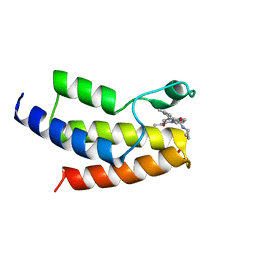 | | Crystal structure of the bromodomain of human BRD9 bound to TP472 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-acetylpyrrolo[1,2-a]pyrimidin-8-yl)-N-cyclopropyl-4-methylbenzamide, Bromodomain-containing protein 9 | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1E
 
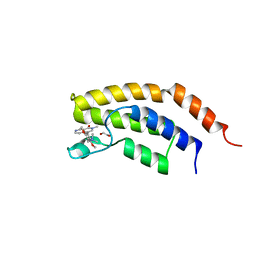 | | Crystal structure of the bromodomain of human BRD7 bound to BI7273 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-[(dimethylamino)methyl]-3,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 7 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6UZF
 
 | | Crystal structure of the unliganded bromodomain of human BRD9 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 9, DIMETHYL SULFOXIDE, ... | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V0U
 
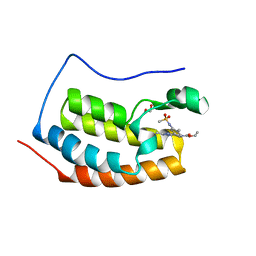 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to bromosporine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Bromosporine | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V17
 
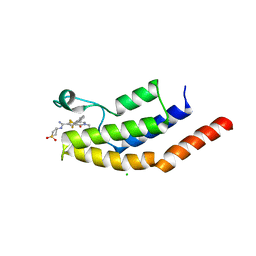 | | Crystal structure of the bromodomain of human BRD7 bound to I-BRD9 | | Descriptor: | Bromodomain-containing protein 7, CHLORIDE ION, N'-[1,1-bis(oxidanylidene)thian-4-yl]-5-ethyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1F
 
 | | Crystal structure of the bromodomain of human BRD7 bound to BI9564 | | Descriptor: | 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 7 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6VGL
 
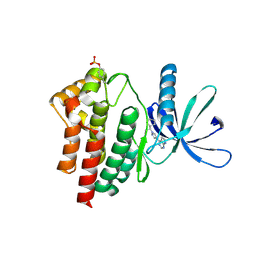 | | JAK2 JH1 in complex with ruxolitinib | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNB
 
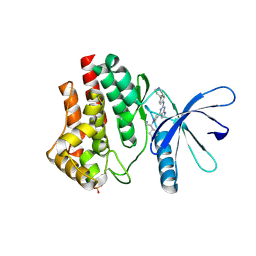 | | JAK2 JH1 in complex with BL2-084 | | Descriptor: | (3S)-3-cyclopentyl-3-[4-(2-{[4-(piperidin-4-yl)phenyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNE
 
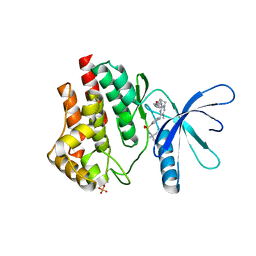 | | JAK2 JH1 in complex with Fedratinib | | Descriptor: | N-tert-butyl-3-{[5-methyl-2-({4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]amino}benzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNC
 
 | | JAK2 JH1 in complex with BL2-096 | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(2-{[4-(piperidin-4-yl)phenyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNG
 
 | | JAK2 JH1 in complex with PN2-118 | | Descriptor: | N-{2-fluoro-5-[(2-{[3-fluoro-4-(1-methylpiperidin-4-yl)phenyl]amino}-5-methylpyrimidin-4-yl)amino]phenyl}-2-methylpropane-2-sulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNH
 
 | | JAK2 JH1 in complex with PN2-123 | | Descriptor: | N-{5-[(2-{[3,5-difluoro-4-(1-methylpiperidin-4-yl)phenyl]amino}-5-methylpyrimidin-4-yl)amino]-2-fluorophenyl}-2-methylpropane-2-sulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNF
 
 | | JAK2 JH1 in complex with MA9-086 | | Descriptor: | N~4~-[1-(tert-butylsulfonyl)-2,3-dihydro-1H-indol-6-yl]-N~2~-[3-fluoro-4-(1-methylpiperidin-4-yl)phenyl]-5-methylpyrimidine-2,4-diamine, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VN8
 
 | | JAK2 JH1 in complex with baricitinib | | Descriptor: | Baricitinib, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
